Directed evolution of broadly crossreactive chemokine-blocking antibodies efficacious in arthritis
- PMID: 29654232
- PMCID: PMC5899157
- DOI: 10.1038/s41467-018-03687-x
Directed evolution of broadly crossreactive chemokine-blocking antibodies efficacious in arthritis
Abstract
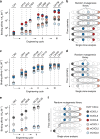
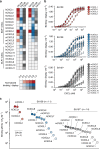
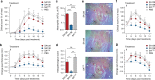
Chemokine receptors typically have multiple ligands. Consequently, treatment with a blocking antibody against a single chemokine is expected to be insufficient for efficacy. Here we show single-chain antibodies can be engineered for broad crossreactivity toward multiple human and mouse proinflammatory ELR+ CXC chemokines. The engineered molecules recognize functional epitopes of ELR+ CXC chemokines and inhibit neutrophil activation ex vivo. Furthermore, an albumin fusion of the most crossreactive single-chain antibody prevents and reverses inflammation in the K/BxN mouse model of arthritis. Thus, we report an approach for the molecular evolution and selection of broadly crossreactive antibodies towards a family of structurally related, yet sequence-diverse protein targets, with general implications for the development of novel therapeutics.
Conflict of interest statement
A.A., A.D.L., and K.D.W. declare that they are named on a provisional patent application 62/546814 entitled “Multiple Specificity Binders of CXC Chemokines and Uses Thereof” that has been filed in the United States Patent and Trademark Office on behalf of the Massachusetts Institute of Technology and the Massachusetts General Hospital. The remaining authors declare no competing interests.
Figures




References
-
- Koch AE, et al. Synovial tissue macrophage as a source of the chemotactic cytokine IL-8. J. Immunol. 1991;147:2187–2195. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

