Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms
- PMID: 29654271
- PMCID: PMC6224406
- DOI: 10.1038/s41375-018-0103-3
Whole-genome sequencing of multiple myeloma reveals oncogenic pathways are targeted somatically through multiple mechanisms
Abstract
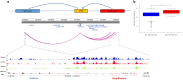
Multiple myeloma (MM) is a biologically heterogeneous malignancy, however, the mechanisms underlying this complexity are incompletely understood. We report an analysis of the whole-genome sequencing of 765 MM patients from CoMMpass. By employing promoter capture Hi-C in naïve B-cells, we identify cis-regulatory elements (CREs) that represent a highly enriched subset of the non-coding genome in which to search for driver mutations. We identify regulatory regions whose mutation significantly alters the expression of genes as candidate non-coding drivers, including copy number variation (CNV) at CREs of MYC and single-nucleotide variants (SNVs) in a PAX5 enhancer. To better inform the interplay between non-coding driver mutations with other driver mechanisms, and their respective roles in oncogenic pathways, we extended our analysis identifying coding drivers in 40 genes, including 11 novel candidates. We demonstrate the same pathways can be targeted by coding and non-coding mutations; exemplified by IRF4 and PRDM1, along with BCL6 and PAX5, genes that are central to plasma cell differentiation. This study reveals new insights into the complex genetic alterations driving MM development and an enhanced understanding of oncogenic pathways.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

