A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus
- PMID: 29654311
- PMCID: PMC5899154
- DOI: 10.1038/s41467-018-03614-0
A TAD boundary is preserved upon deletion of the CTCF-rich Firre locus
Abstract
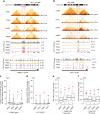
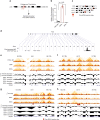
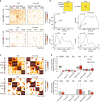
The binding of the transcriptional regulator CTCF to the genome has been implicated in the formation of topologically associated domains (TADs). However, the general mechanisms of folding the genome into TADs are not fully understood. Here we test the effects of deleting a CTCF-rich locus on TAD boundary formation. Using genome-wide chromosome conformation capture (Hi-C), we focus on one TAD boundary on chromosome X harboring ~ 15 CTCF binding sites and located at the long non-coding RNA (lncRNA) locus Firre. Specifically, this TAD boundary is invariant across evolution, tissues, and temporal dynamics of X-chromosome inactivation. We demonstrate that neither the deletion of this locus nor the ectopic insertion of Firre cDNA or its ectopic expression are sufficient to alter TADs in a sex-specific or allele-specific manner. In contrast, Firre's deletion disrupts the chromatin super-loop formation of the inactive X-chromosome. Collectively, our findings suggest that apart from CTCF binding, additional mechanisms may play roles in establishing TAD boundary formation.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

