Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs
- PMID: 29654314
- PMCID: PMC5899086
- DOI: 10.1038/s41598-018-24289-z
Comparative biogeography of the gut microbiome between Jinhua and Landrace pigs
Abstract
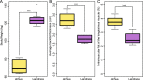
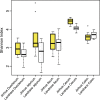
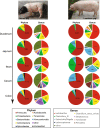
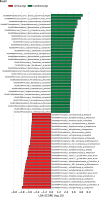
The intestinal microbiome is critically important in shaping a variety of host physiological responses. However, it remains elusive on how gut microbiota impacts overall growth and more specifically, adipogenesis. Using the pig as an animal model, we compared the differences in bacterial community structure throughout the intestinal tract in two breeds (Landrace and Jinhua) of pigs with distinct phenotypes. The Landrace is a commercial purebred and the Jinhua is a Chinese indigenous, slow-growing breed with high propensity for fat deposition. Using 16S rRNA gene sequencing, we revealed that the bacterial communities are more diverse in the duodenum, jejunum, and cecum of Jinhua pigs than in those of Landrace pigs, whereas the ileal and colonic microbiota show a similar complexity between the two breeds. Furthermore, a number of bacterial taxa differentially exist in Jinhua and Landrace pigs throughout the entire intestinal tract, with the jejunal and ileal microbiome showing the greatest contrast. Functional prediction of the bacterial community suggested increased fatty acid biosynthesis in Jinghua pigs, which could partially explain their adiposity phenotype. Further studies are warranted to experimentally verify the relative contribution of each enriched bacterial species and their effect on adipogenesis and animal growth.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Core gut microbiota in Jinhua pigs and its correlation with strain, farm and weaning age.J Microbiol. 2018 May;56(5):346-355. doi: 10.1007/s12275-018-7486-8. Epub 2018 May 2. J Microbiol. 2018. PMID: 29721832
-
Changes in gut microbial populations, intestinal morphology, expression of tight junction proteins, and cytokine production between two pig breeds after challenge with Escherichia coli K88: a comparative study.J Anim Sci. 2013 Dec;91(12):5614-25. doi: 10.2527/jas.2013-6528. Epub 2013 Oct 14. J Anim Sci. 2013. PMID: 24126267
-
Uncovering the composition of microbial community structure and metagenomics among three gut locations in pigs with distinct fatness.Sci Rep. 2016 Jun 3;6:27427. doi: 10.1038/srep27427. Sci Rep. 2016. PMID: 27255518 Free PMC article.
-
The pig gut microbial diversity: Understanding the pig gut microbial ecology through the next generation high throughput sequencing.Vet Microbiol. 2015 Jun 12;177(3-4):242-51. doi: 10.1016/j.vetmic.2015.03.014. Epub 2015 Mar 23. Vet Microbiol. 2015. PMID: 25843944 Review.
-
The significance of the gut microbiome in children with functional constipation.Adv Clin Exp Med. 2021 Apr;30(4):471-480. doi: 10.17219/acem/131215. Adv Clin Exp Med. 2021. PMID: 33908196 Review.
Cited by
-
Gut Microbiota Is a Major Contributor to Adiposity in Pigs.Front Microbiol. 2018 Dec 10;9:3045. doi: 10.3389/fmicb.2018.03045. eCollection 2018. Front Microbiol. 2018. PMID: 30619136 Free PMC article.
-
Fecal microbiota transplantation in pigs: current status and future perspective.Anim Microbiome. 2025 Jul 20;7(1):76. doi: 10.1186/s42523-025-00440-w. Anim Microbiome. 2025. PMID: 40685369 Free PMC article. Review.
-
Ileal Microbiota Alters the Immunity Statues to Affect Body Weight in Muscovy Ducks.Front Immunol. 2022 Feb 10;13:844102. doi: 10.3389/fimmu.2022.844102. eCollection 2022. Front Immunol. 2022. PMID: 35222437 Free PMC article.
-
The Role of Gut Microbiota in the Skeletal Muscle Development and Fat Deposition in Pigs.Antibiotics (Basel). 2022 Jun 11;11(6):793. doi: 10.3390/antibiotics11060793. Antibiotics (Basel). 2022. PMID: 35740199 Free PMC article.
-
A Comprehensive Analysis of the Colonic Flora Diversity, Short Chain Fatty Acid Metabolism, Transcripts, and Biochemical Indexes in Heat-Stressed Pigs.Front Immunol. 2021 Oct 21;12:717723. doi: 10.3389/fimmu.2021.717723. eCollection 2021. Front Immunol. 2021. PMID: 34745096 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

