A 360° view of circular RNAs: From biogenesis to functions
- PMID: 29655315
- PMCID: PMC6002912
- DOI: 10.1002/wrna.1478
A 360° view of circular RNAs: From biogenesis to functions
Abstract
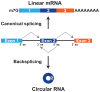
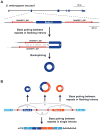
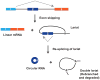
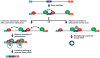
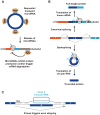
The first circular RNA (circRNA) was identified more than 40 years ago, but it was only recently appreciated that circRNAs are common outputs of many eukaryotic protein-coding genes. Some circRNAs accumulate to higher levels than their associated linear mRNAs, especially in the nervous system, and have clear regulatory functions that result in organismal phenotypes. The pre-mRNA splicing machinery generates circRNAs via backsplicing reactions, which are often facilitated by intronic repeat sequences that base pair to one another and bring the intervening splice sites into close proximity. When spliceosomal components are limiting, circRNAs can become the preferred gene output, and backsplicing reactions are further controlled by exon skipping events and the combinatorial action of RNA binding proteins. This allows circRNAs to be expressed in a tissue- and stage-specific manner. Once generated, circRNAs are highly stable transcripts that often accumulate in the cytoplasm. The functions of most circRNAs remain unknown, but some can regulate the activities of microRNAs or be translated to produce proteins. Circular RNAs can further interface with the immune system as well as control gene expression events in the nucleus, including alternative splicing decisions. Circular RNAs thus represent a large class of RNA molecules that are tightly regulated, and it is becoming increasingly clear that they likely impact many biological processes. This article is categorized under: RNA Processing > Splicing Mechanisms RNA Structure and Dynamics > Influence of RNA Structure in Biological Systems RNA Evolution and Genomics > RNA and Ribonucleoprotein Evolution RNA Evolution and Genomics > Computational Analyses of RNA.
Keywords: CDR1as; Laccase2; R-loop; alternative splicing; backsplicing; circRNA; exon skipping; immune surveillance; microRNA; noncoding RNA; pre-mRNA splicing; scrambled exons; spliceosome; translation; transposable elements.
© 2018 Wiley Periodicals, Inc.
Conflict of interest statement
The author has declared no conflicts of interest for this article.
Figures
References
-
- Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. - PubMed
-
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL. Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell. 2000;101:671–684. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

