Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium
- PMID: 29657097
- PMCID: PMC5995784
- DOI: 10.1016/j.stemcr.2018.03.013
Single-Cell Transcriptomic Profiling of Pluripotent Stem Cell-Derived SCGB3A2+ Airway Epithelium
Abstract
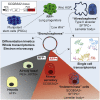
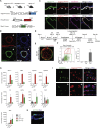
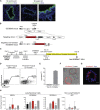
Lung epithelial lineages have been difficult to maintain in pure form in vitro, and lineage-specific reporters have proven invaluable for monitoring their emergence from cultured pluripotent stem cells (PSCs). However, reporter constructs for tracking proximal airway lineages generated from PSCs have not been previously available, limiting the characterization of these cells. Here, we engineer mouse and human PSC lines carrying airway secretory lineage reporters that facilitate the tracking, purification, and profiling of this lung subtype. Through bulk and single-cell-based global transcriptomic profiling, we find PSC-derived airway secretory cells are susceptible to phenotypic plasticity exemplified by the tendency to co-express both a proximal airway secretory program as well as an alveolar type 2 cell program, which can be minimized by inhibiting endogenous Wnt signaling. Our results provide global profiles of engineered lung cell fates, a guide for improving their directed differentiation, and a human model of the developing airway.
Keywords: airway; alveoli; directed differentiation; lung epithelium; pluripotent stem cells; single-cell RNA sequencing.
Copyright © 2018 The Author(s). Published by Elsevier Inc. All rights reserved.
Figures





References
-
- Boers J.E., Ambergen A.W., Thunnissen F.B. Number and proliferation of Clara cells in normal human airway epithelium. Am. J. Respir. Crit. Care Med. 1999;159(5 Pt 1):1585–1591. - PubMed
-
- Brasch F., Johnen G., Winn-Brasch A., Guttentag S.H., Schmiedl A., Kapp N., Suzuki Y., Müller K.M., Richter J., Hawgood S. Surfactant protein B in type II pneumocytes and intra-alveolar surfactant forms of human lungs. Am. J. Respir. Cell Mol. Biol. 2004;30:449–458. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 HL128172/HL/NHLBI NIH HHS/United States
- U01 TR001810/TR/NCATS NIH HHS/United States
- R24 HL123828/HL/NHLBI NIH HHS/United States
- T32 HL007035/HL/NHLBI NIH HHS/United States
- U01 HL134745/HL/NHLBI NIH HHS/United States
- R01 HL059959/HL/NHLBI NIH HHS/United States
- F31 HL129777/HL/NHLBI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- R01 HL095993/HL/NHLBI NIH HHS/United States
- F31 HL134274/HL/NHLBI NIH HHS/United States
- U01 HL134766/HL/NHLBI NIH HHS/United States
- R01 HL122442/HL/NHLBI NIH HHS/United States
- UL1 TR001430/TR/NCATS NIH HHS/United States
- U01 HL099997/HL/NHLBI NIH HHS/United States
- R01 GM108807/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

