RNA Biomarkers: Frontier of Precision Medicine for Cancer
- PMID: 29657281
- PMCID: PMC5832009
- DOI: 10.3390/ncrna3010009
RNA Biomarkers: Frontier of Precision Medicine for Cancer
Abstract
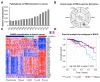
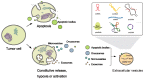
As an essential part of central dogma, RNA delivers genetic and regulatory information and reflects cellular states. Based on high-throughput sequencing technologies, cumulating data show that various RNA molecules are able to serve as biomarkers for the diagnosis and prognosis of various diseases, for instance, cancer. In particular, detectable in various bio-fluids, such as serum, saliva and urine, extracellular RNAs (exRNAs) are emerging as non-invasive biomarkers for earlier cancer diagnosis, tumor progression monitor, and prediction of therapy response. In this review, we summarize the latest studies on various types of RNA biomarkers, especially extracellular RNAs, in cancer diagnosis and prognosis, and illustrate several well-known RNA biomarkers of clinical utility. In addition, we describe and discuss general procedures and issues in investigating exRNA biomarkers, and perspectives on utility of exRNAs in precision medicine.
Keywords: biomarker; cancer; exRNA; precision medicine.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

