Computational Characterization of ncRNA Fragments in Various Tissues of the Brassica rapa Plant
- PMID: 29657288
- PMCID: PMC5831936
- DOI: 10.3390/ncrna3020017
Computational Characterization of ncRNA Fragments in Various Tissues of the Brassica rapa Plant
Abstract
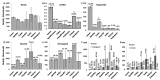
Recently, a novel type of non-coding RNA (ncRNA), known as ncRNA fragments or ncRFs, has been characterised in various organisms, including plants. The biogenesis mechanism, function and abundance of ncRFs stemming from various ncRNAs are poorly understood, especially in plants. In this work, we have computationally analysed the composition of ncRNAs and the fragments that derive from them in various tissues of Brassica rapa plants, including leaves, meristem tissue, pollen, unfertilized and fertilized ova, embryo and endosperm. Detailed analysis of transfer RNA (tRNA) fragments (tRFs), ribosomal RNA (rRNA) fragments (rRFs), small nucleolar RNA (snoRNA) fragments (snoRFs) and small nuclear RNA (snRNA) fragments (snRFs) showed a predominance of tRFs, with the 26 nucleotides (nt) fraction being the largest. Mapping ncRF reads to full-length mature ncRNAs showed a strong bias for one or both termini. tRFs mapped predominantly to the 5' end, whereas snRFs mapped to the 3' end, suggesting that there may be specific biogenesis and retention mechanisms. In the case of tRFs, specific isoacceptors were enriched, including tRNAGly(UCC) and tRFAsp(GUC). The analysis showed that the processing of 26-nt tRF5' occurred by cleavage at the last unpaired nucleotide of the loop between the D arm and the anticodon arm. Further support for the functionality of ncRFs comes from the analysis of binding between ncRFs and their potential targets. A higher average percentage of binding at the first half of fragments was observed, with the highest percentage being at 2-6 nt. To summarise, our analysis showed that ncRFs in B. rapa are abundantly produced in a tissue-specific manner, with bias toward a terminus, the bias toward the size of generated fragments and the bias toward the targeting of specific biological processes.
Keywords: Brassica rapa; apical meristem; embryo; endosperm; leaves; ncRFs; ncRNA; ncRNA fragments; non-coding RNA; pollen; pollinated ovules; rRF; snRF; snoRF; tRF; unpollinated ovules.
Conflict of interest statement
The authors declare no conflict of interest. The founding sponsors had no role in the design of the study, in the collection, analyses, and interpretation of data, in the writing of the manuscript, or in the decision to publish the results.
Figures








References
-
- Yang J.X., Rastetter R.H., Wilhelm D. Non-coding RNAs: An Introduction. Adv. Exp. Med. Biol. 2016;886:13–32. - PubMed
-
- Garcia-Silva M.R., Frugier M., Tosar J.P., Correa-Dominguez A., Ronalte-Alves L., Parodi-Talice A., Rovira C., Robello C., Goldenberg S., Cayota A. A population of tRNA-derived small RNAs is actively produced in Trypanosoma cruzi and recruited to specific cytoplasmic granules. Mol. Biochem. Parasitol. 2010;171:64–73. doi: 10.1016/j.molbiopara.2010.02.003. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

