Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription
- PMID: 29659567
- PMCID: PMC5919686
- DOI: 10.1371/journal.pgen.1007351
Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription
Abstract
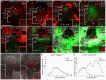
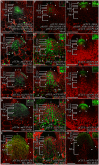
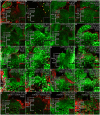
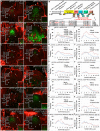
Concentration-dependent transcriptional regulation and the spatial regulation of transcription factor levels are poorly studied in plant development. WUSCHEL, a stem cell-promoting homeodomain transcription factor, accumulates at a higher level in the rib meristem than in the overlying central zone, which harbors stem cells in the shoot apical meristems of Arabidopsis thaliana. The differential accumulation of WUSCHEL in adjacent cells is critical for the spatial regulation and levels of CLAVATA3, a negative regulator of WUSCHEL transcription. Earlier studies have revealed that DNA-dependent dimerization, subcellular partitioning and protein destabilization control WUSCHEL protein levels and spatial accumulation. Moreover, the destabilization of WUSCHEL may also depend on the protein concentration. However, the roles of extrinsic spatial cues in maintaining differential accumulation of WUS are not understood. Through transient manipulation of hormone levels, hormone response patterns and analysis of the receptor mutants, we show that cytokinin signaling in the rib meristem acts through the transcriptional regulatory domains, the acidic domain and the WUSCHEL-box, to stabilize the WUS protein. Furthermore, we show that the same WUSCHEL-box functions as a degron sequence in cytokinin deficient regions in the central zone, leading to the destabilization of WUSCHEL. The coupled functions of the WUSCHEL-box in nuclear retention as described earlier, together with cytokinin sensing, reinforce higher nuclear accumulation of WUSCHEL in the rib meristem. In contrast a sub-threshold level may expose the WUSCHEL-box to destabilizing signals in the central zone. Thus, the cytokinin signaling acts as an asymmetric spatial cue in stabilizing the WUSCHEL protein to lead to its differential accumulation in neighboring cells, which is critical for concentration-dependent spatial regulation of CLAVATA3 transcription and meristem maintenance. Furthermore, our work shows that cytokinin response is regulated independently of the WUSCHEL function which may provide robustness to the regulation of WUSCHEL concentration.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Rushlow CA, Han K, Manley JL, Levine M. The graded distribution of the dorsal morphogen is initiated by selective nuclear transport in Drosophila. Cell. 1989;59: 1165–1177. - PubMed
-
- Nellen D, Burke R, Struhl G, Basler K. Direct and long-range action of a DPP morphogen gradient. Cell. 1996;85: 357–368. - PubMed
-
- Briscoe J, Ericson J. The specification of neuronal identity by graded Sonic Hedgehog signalling. Semin Cell Dev Biol. 1999;10: 353–362. doi: 10.1006/scdb.1999.0295 - DOI - PubMed
-
- Steeves TA, Sussex IM. Patterns in plant development. Cambridge University Press; 1989.
-
- Reddy GV, Heisler MG, Ehrhardt DW, Meyerowitz EM. Real-time lineage analysis reveals oriented cell divisions associated with morphogenesis at the shoot apex of Arabidopsis thaliana. Development. 2004;131: 4225–4237. doi: 10.1242/dev.01261 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

