N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection
- PMID: 29659627
- PMCID: PMC5919695
- DOI: 10.1371/journal.ppat.1006995
N6-methyladenosine modification and the YTHDF2 reader protein play cell type specific roles in lytic viral gene expression during Kaposi's sarcoma-associated herpesvirus infection
Abstract
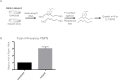
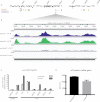
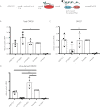
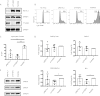
Methylation at the N6 position of adenosine (m6A) is a highly prevalent and reversible modification within eukaryotic mRNAs that has been linked to many stages of RNA processing and fate. Recent studies suggest that m6A deposition and proteins involved in the m6A pathway play a diverse set of roles in either restricting or modulating the lifecycles of select viruses. Here, we report that m6A levels are significantly increased in cells infected with the oncogenic human DNA virus Kaposi's sarcoma-associated herpesvirus (KSHV). Transcriptome-wide m6A-sequencing of the KSHV-positive renal carcinoma cell line iSLK.219 during lytic reactivation revealed the presence of m6A across multiple kinetic classes of viral transcripts, and a concomitant decrease in m6A levels across much of the host transcriptome. However, we found that depletion of the m6A machinery had differential pro- and anti-viral impacts on viral gene expression depending on the cell-type analyzed. In iSLK.219 and iSLK.BAC16 cells the pathway functioned in a pro-viral manner, as depletion of the m6A writer METTL3 and the reader YTHDF2 significantly impaired virion production. In iSLK.219 cells the defect was linked to their roles in the post-transcriptional accumulation of the major viral lytic transactivator ORF50, which is m6A modified. In contrast, although the ORF50 mRNA was also m6A modified in KSHV infected B cells, ORF50 protein expression was instead increased upon depletion of METTL3, or, to a lesser extent, YTHDF2. These results highlight that the m6A pathway is centrally involved in regulating KSHV gene expression, and underscore how the outcome of this dynamically regulated modification can vary significantly between cell types.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
Kaposi's Sarcoma-Associated Herpesvirus Lytic Replication Is Independent of Anaphase-Promoting Complex Activity.J Virol. 2020 Jun 16;94(13):e02079-19. doi: 10.1128/JVI.02079-19. Print 2020 Jun 16. J Virol. 2020. PMID: 32295923 Free PMC article.
-
Identification of Novel Kaposi's Sarcoma-Associated Herpesvirus Orf50 Transcripts: Discovery of New RTA Isoforms with Variable Transactivation Potential.J Virol. 2016 Dec 16;91(1):e01434-16. doi: 10.1128/JVI.01434-16. Print 2017 Jan 1. J Virol. 2016. PMID: 27795414 Free PMC article.
-
Effects of the NEDD8-Activating Enzyme Inhibitor MLN4924 on Lytic Reactivation of Kaposi's Sarcoma-Associated Herpesvirus.J Virol. 2017 Sep 12;91(19):e00505-17. doi: 10.1128/JVI.00505-17. Print 2017 Oct 1. J Virol. 2017. PMID: 28701396 Free PMC article.
-
The RNA Epitranscriptome of DNA Viruses.J Virol. 2018 Oct 29;92(22):e00696-18. doi: 10.1128/JVI.00696-18. Print 2018 Nov 15. J Virol. 2018. PMID: 30185592 Free PMC article. Review.
-
RNA epitranscriptomics: Regulation of infection of RNA and DNA viruses by N6 -methyladenosine (m6 A).Rev Med Virol. 2018 Jul;28(4):e1983. doi: 10.1002/rmv.1983. Epub 2018 Apr 26. Rev Med Virol. 2018. PMID: 29698584 Free PMC article. Review.
Cited by
-
The m6A epitranscriptome opens a new charter in immune system logic.Epigenetics. 2021 Aug;16(8):819-837. doi: 10.1080/15592294.2020.1827722. Epub 2020 Oct 19. Epigenetics. 2021. PMID: 33070685 Free PMC article.
-
Viral RNA N6-methyladenosine modification modulates both innate and adaptive immune responses of human respiratory syncytial virus.PLoS Pathog. 2021 Dec 20;17(12):e1010142. doi: 10.1371/journal.ppat.1010142. eCollection 2021 Dec. PLoS Pathog. 2021. PMID: 34929018 Free PMC article.
-
The impact of RNA modifications on the biology of DNA virus infection.Eur J Cell Biol. 2022 Jun-Aug;101(3):151239. doi: 10.1016/j.ejcb.2022.151239. Epub 2022 May 21. Eur J Cell Biol. 2022. PMID: 35623231 Free PMC article. Review.
-
The roles and mechanisms of YTH domain-containing proteins in cancer development and progression.Am J Cancer Res. 2020 Apr 1;10(4):1068-1084. eCollection 2020. Am J Cancer Res. 2020. PMID: 32368386 Free PMC article. Review.
-
Epigenetic and epitranscriptomic regulation during oncogenic γ-herpesvirus infection.Front Microbiol. 2025 Jan 7;15:1484455. doi: 10.3389/fmicb.2024.1484455. eCollection 2024. Front Microbiol. 2025. PMID: 39839102 Free PMC article. Review.
References
-
- Roundtree IA, Evans ME, Pan T, He C. Dynamic RNA Modifications in Gene Expression Regulation. Cell. 2017;169: 1187–1200. doi: 10.1016/j.cell.2017.05.045 - DOI - PMC - PubMed
-
- Wang X, Lu Z, Gomez A, Hon GC, Yue Y, Han D, et al. N(6)-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2013;505: 117–120. doi: 10.1038/nature12730 - DOI - PMC - PubMed
-
- Wang X, Zhao BS, Roundtree IA, Lu Z, Han D, Ma H, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161: 1388–1399. doi: 10.1016/j.cell.2015.05.014 - DOI - PMC - PubMed
-
- Fu Y, Dominissini D, Rechavi G, He C. Gene expression regulation mediated through reversible m6A RNA methylation. Nat Rev Genet. 2014;15: 293–306. doi: 10.1038/nrg3724 - DOI - PubMed
-
- Zhao BS, Wang X, Beadell AV, Lu Z, Shi H, Kuuspalu A, et al. m(6)A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542: 475–478. doi: 10.1038/nature21355 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

