Biophysical characterization of actin bundles generated by the Chlamydia trachomatis Tarp effector
- PMID: 29660331
- PMCID: PMC5928783
- DOI: 10.1016/j.bbrc.2018.04.093
Biophysical characterization of actin bundles generated by the Chlamydia trachomatis Tarp effector
Abstract
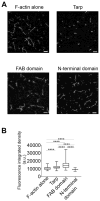
Chlamydia trachomatis entry into host cells is mediated by pathogen-directed remodeling of the actin cytoskeleton. The chlamydial type III secreted effector, translocated actin recruiting phosphoprotein (Tarp), has been implicated in the recruitment of actin to the site of internalization. Tarp harbors G-actin binding and proline rich domains required for Tarp-mediated actin nucleation as well as unique F-actin binding domains implicated in the formation of actin bundles. Little is known about the mechanical properties of actin bundles generated by Tarp or the mechanism by which Tarp mediates actin bundle formation. In order to characterize the actin bundles and elucidate the role of different Tarp domains in the bundling process, purified Tarp effectors and Tarp truncation mutants were analyzed using Total Internal Reflection Fluorescence (TIRF) microscopy. Our data indicate that Tarp mediated actin bundling is independent of actin nucleation and the F-actin binding domains are sufficient to bundle actin filaments. Additionally, Tarp-mediated actin bundles demonstrate distinct bending stiffness compared to those crosslinked by the well characterized actin bundling proteins fascin and alpha-actinin, suggesting Tarp may employ a novel actin bundling strategy. The capacity of the Tarp effector to generate novel actin bundles likely contributes to chlamydia's efficient mechanism of entry into human cells.
Keywords: Actin bundles; Bending persistence length; Chlamydia trachomatis; Cytoskeleton; Effector; Tarp.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

