Risk factors and outcomes for the Q151M and T69 insertion HIV-1 resistance mutations in historic UK data
- PMID: 29661246
- PMCID: PMC5902836
- DOI: 10.1186/s12981-018-0198-7
Risk factors and outcomes for the Q151M and T69 insertion HIV-1 resistance mutations in historic UK data
Abstract
Background: The prevalence of HIV-1 resistance to antiretroviral therapies (ART) has declined in high-income countries over recent years, but drug resistance remains a substantial concern in many low and middle-income countries. The Q151M and T69 insertion (T69i) resistance mutations in the viral reverse transcriptase gene can reduce susceptibility to all nucleoside/tide analogue reverse transcriptase inhibitors, motivating the present study to investigate the risk factors and outcomes associated with these mutations.
Methods: We considered all data in the UK HIV Drug Resistance Database for blood samples obtained in the period 1997-2014. Where available, treatment history and patient outcomes were obtained through linkage to the UK Collaborative HIV Cohort study. A matched case-control approach was used to assess risk factors associated with the appearance of each of the mutations in ART-experienced patients, and survival analysis was used to investigate factors associated with viral suppression. A further analysis using matched controls was performed to investigate the impact of each mutation on survival.
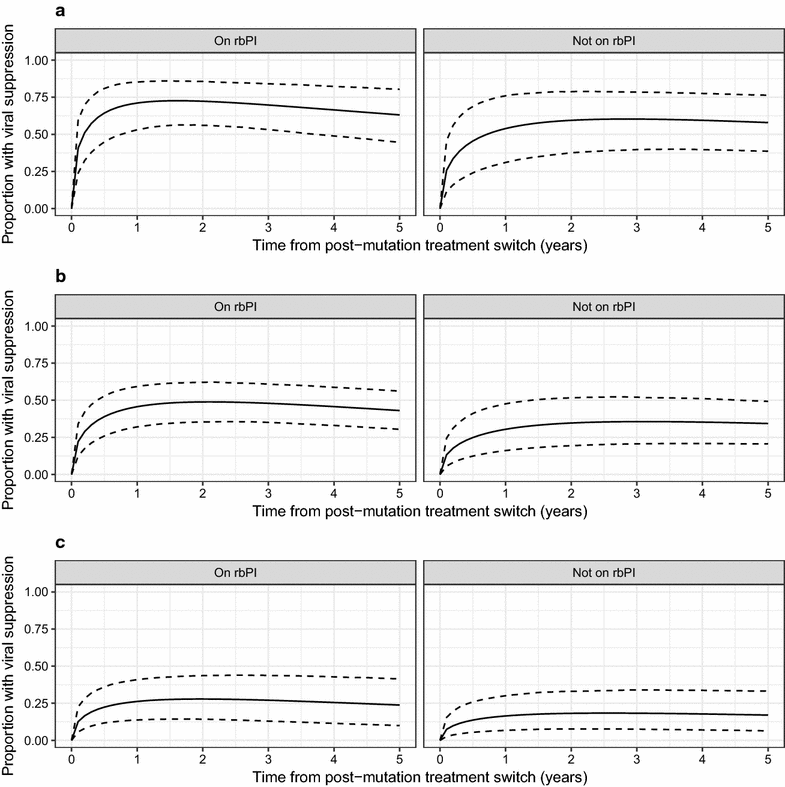
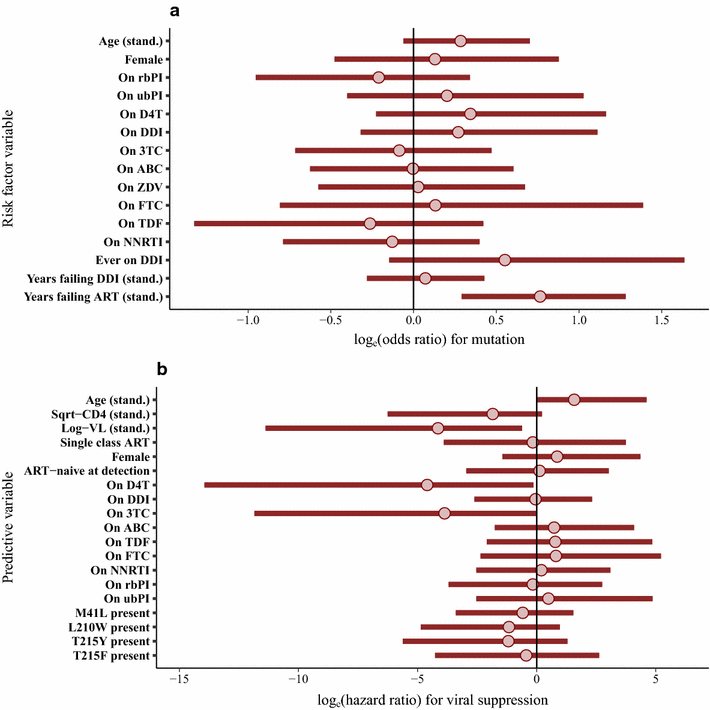
Results: A total of 180 patients with Q151M mutation and 85 with T69i mutation were identified, almost entirely from before 2006. Occurrence of both the Q151M and T69i mutations was strongly associated with cumulative period of virological failure while on ART, and for Q151M there was a particular positive association with use of stavudine and negative association with use of boosted-protease inhibitors. Subsequent viral suppression was negatively associated with viral load at sequencing for both mutations, and for Q151M we found a negative association with didanosine use but a positive association with boosted-protease inhibitor use. The results obtained in these analyses were also consistent with potentially large associations with other drugs. Analyses were inconclusive regarding associations between the mutations and mortality, but mortality was high for patients with low CD4 at detection.
Conclusions: The Q151M and T69i resistance mutations are now very rare in the UK. Our results suggest that good outcomes are possible for people with these mutations. However, in this historic sample, viral load and CD4 at detection were important factors in determining prognosis.
Keywords: 151 complex; 69 insertion complex; HIV; Multi-NRTI resistance; Multidrug resistance; NRTI.
Figures



References
-
- UK HIV Drug Resistance Database. HIV drug resistance in the UK. 2017. http://www.hivrdb.org.uk/hiv-drug-resistance-uk. Accessed 13 Sept 2017.
-
- Ssemwanga D, Lihana RW, Ugoji C, Al Abimiku, Nkengasong J, Dakum P, et al. Update on HIV-1 acquired and transmitted drug resistance in Africa. AIDS Rev. 2015;17:3–20. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

