In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design
- PMID: 29662055
- PMCID: PMC5902452
- DOI: 10.1038/s41467-018-03746-3
In silico optimization of a guava antimicrobial peptide enables combinatorial exploration for peptide design
Abstract
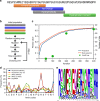
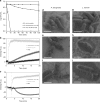
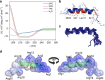
Plants are extensively used in traditional medicine, and several plant antimicrobial peptides have been described as potential alternatives to conventional antibiotics. However, after more than four decades of research no plant antimicrobial peptide is currently used for treating bacterial infections, due to their length, post-translational modifications or high dose requirement for a therapeutic effect . Here we report the design of antimicrobial peptides derived from a guava glycine-rich peptide using a genetic algorithm. This approach yields guavanin peptides, arginine-rich α-helical peptides that possess an unusual hydrophobic counterpart mainly composed of tyrosine residues. Guavanin 2 is characterized as a prototype peptide in terms of structure and activity. Nuclear magnetic resonance analysis indicates that the peptide adopts an α-helical structure in hydrophobic environments. Guavanin 2 is bactericidal at low concentrations, causing membrane disruption and triggering hyperpolarization. This computational approach for the exploration of natural products could be used to design effective peptide antibiotics.
Conflict of interest statement
The authors declare no competing interests.
Figures

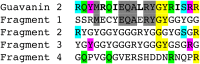
References
-
- Cândido, E. S. et al. in Science Against Microbial Pathogens: Communicating Current Research and Technological Advances (ed. Méndez-Vilas, A.) 951–960 (Formatex, Badajoz, Spain, 2011).
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

