Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity
- PMID: 29662164
- PMCID: PMC6022759
- DOI: 10.1038/s41588-018-0102-3
Transcription factors operate across disease loci, with EBNA2 implicated in autoimmunity
Abstract
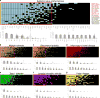
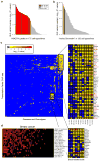
Explaining the genetics of many diseases is challenging because most associations localize to incompletely characterized regulatory regions. Using new computational methods, we show that transcription factors (TFs) occupy multiple loci associated with individual complex genetic disorders. Application to 213 phenotypes and 1,544 TF binding datasets identified 2,264 relationships between hundreds of TFs and 94 phenotypes, including androgen receptor in prostate cancer and GATA3 in breast cancer. Strikingly, nearly half of systemic lupus erythematosus risk loci are occupied by the Epstein-Barr virus EBNA2 protein and many coclustering human TFs, showing gene-environment interaction. Similar EBNA2-anchored associations exist in multiple sclerosis, rheumatoid arthritis, inflammatory bowel disease, type 1 diabetes, juvenile idiopathic arthritis and celiac disease. Instances of allele-dependent DNA binding and downstream effects on gene expression at plausibly causal variants support genetic mechanisms dependent on EBNA2. Our results nominate mechanisms that operate across risk loci within disease phenotypes, suggesting new models for disease origins.
Conflict of interest statement
J.B.H., M.T.W., and L.C.K. have a submitted patent application relating to these findings. A.B. is a co-founder of Datirium, LLC.
Figures


Comment in
-
Role of Epstein-Barr virus infection in SLE: gene-environment interactions at the molecular level.Ann Rheum Dis. 2018 Sep;77(9):1249-1250. doi: 10.1136/annrheumdis-2018-213783. Epub 2018 Jun 1. Ann Rheum Dis. 2018. PMID: 29858172 No abstract available.
-
A role for the Epstein-Barr virus in multiple sclerosis aetiology?J Neurol. 2022 Jul;269(7):3962-3963. doi: 10.1007/s00415-022-11177-w. Epub 2022 May 31. J Neurol. 2022. PMID: 35639199 Free PMC article. No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
- U01 AI130830/AI/NIAID NIH HHS/United States
- R01 NS099068/NS/NINDS NIH HHS/United States
- KL2 TR001426/TR/NCATS NIH HHS/United States
- R01 DK107502/DK/NIDDK NIH HHS/United States
- P30 DK078392/DK/NIDDK NIH HHS/United States
- R24 HL105333/HL/NHLBI NIH HHS/United States
- P30 AR070549/AR/NIAMS NIH HHS/United States
- DP2 GM119134/GM/NIGMS NIH HHS/United States
- R01 AI031584/AI/NIAID NIH HHS/United States
- R21 HG008186/HG/NHGRI NIH HHS/United States
- R01 AI024717/AI/NIAID NIH HHS/United States
- U01 HG008666/HG/NHGRI NIH HHS/United States
- UL1 TR001425/TR/NCATS NIH HHS/United States
- I01 BX001834/BX/BLRD VA/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

