Dynamic coupling between conformations and nucleotide states in DNA gyrase
- PMID: 29662209
- PMCID: PMC10121156
- DOI: 10.1038/s41589-018-0037-0
Dynamic coupling between conformations and nucleotide states in DNA gyrase
Abstract
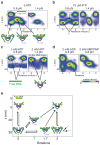
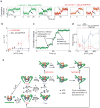
Gyrase is an essential bacterial molecular motor that supercoils DNA using a conformational cycle in which chiral wrapping of > 100 base pairs confers directionality on topoisomerization. To understand the mechanism of this nucleoprotein machine, global structural transitions must be mapped onto the nucleotide cycle of ATP binding, hydrolysis and product release. Here we investigate coupling mechanisms using single-molecule tracking of DNA rotation and contraction during Escherichia coli gyrase activity under varying nucleotide conditions. We find that ADP must be exchanged for ATP to drive the rate-limiting remodeling transition that generates the chiral wrap. ATP hydrolysis accelerates subsequent duplex strand passage and is required for resetting the enzyme and recapturing transiently released DNA. Our measurements suggest how gyrase coordinates DNA rearrangements with the dynamics of its ATP-driven protein gate, how the motor minimizes futile cycles of ATP hydrolysis and how gyrase may respond to changing cellular energy levels to link gene expression with metabolism.
Conflict of interest statement
The authors declare no competing financial interests.
Figures



References
-
- Dutta R, Inouye M. GHKL, an emergent ATPase/kinase superfamily. Trends in biochemical sciences. 2000;25:24–8. - PubMed
-
- Saraste M, Sibbald PR, Wittinghofer A. The P-loop--a common motif in ATP- and GTP-binding proteins. Trends in biochemical sciences. 1990;15:430–4. - PubMed
-
- Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. - PubMed
-
- Wang JC. Cellular roles of DNA topoisomerases: a molecular perspective. Nature reviews Molecular cell biology. 2002;3:430–40. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

