Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance
- PMID: 29662238
- PMCID: PMC6252060
- DOI: 10.1038/s41568-018-0001-z
Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance
Erratum in
-
Publisher Correction: Structural underpinnings of oestrogen receptor mutations in endocrine therapy resistance.Nat Rev Cancer. 2018 Oct;18(10):662. doi: 10.1038/s41568-018-0053-0. Nat Rev Cancer. 2018. PMID: 30185950
Abstract
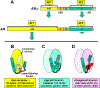
Oestrogen receptor-α (ERα), a key driver of breast cancer, normally requires oestrogen for activation. Mutations that constitutively activate ERα without the need for hormone binding are frequently found in endocrine-therapy-resistant breast cancer metastases and are associated with poor patient outcomes. The location of these mutations in the ER ligand-binding domain and their impact on receptor conformation suggest that they subvert distinct mechanisms that normally maintain the low basal state of wild-type ERα in the absence of hormone. Such mutations provide opportunities to probe fundamental issues underlying ligand-mediated control of ERα activity. Instructive contrasts between these ERα mutations and those that arise in the androgen receptor (AR) during anti-androgen treatment of prostate cancer highlight differences in how activation functions in ERs and AR control receptor activity, how hormonal pressures (deprivation versus antagonism) drive the selection of phenotypically different mutants, how altered protein conformations can reduce antagonist potency and how altered ligand-receptor contacts can invert the response that a receptor has to an agonist ligand versus an antagonist ligand. A deeper understanding of how ligand regulation of receptor conformation is linked to receptor function offers a conceptual framework for developing new anti-oestrogens that might be more effective in preventing and treating breast cancer.
Figures


References
-
- Katzenellenbogen BS & Frasor J Therapeutic targeting in the estrogen receptor hormonal pathway. Semin. Oncol 31, 28–38, (2004). - PubMed
-
- Rugo HS et al. Endocrine Therapy for Hormone Receptor-Positive Metastatic Breast Cancer: American Society of Clinical Oncology Guideline. Journal of clinical oncology : official journal of the American Society of Clinical Oncology 34, 3069–3103, (2016). - PubMed
-
- Tryfonidis K, Zardavas D, Katzenellenbogen BS & Piccart M Endocrine treatment in breast cancer: Cure, resistance and beyond. Cancer treatment reviews 50, 68–81, (2016). - PubMed
-
- Pratt WB & Toft DO Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery. Exp Biol Med (Maywood) 228, 111–133, (2003). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

