Prediction of Cacao (Theobroma cacao) Resistance to Moniliophthora spp. Diseases via Genome-Wide Association Analysis and Genomic Selection
- PMID: 29662497
- PMCID: PMC5890178
- DOI: 10.3389/fpls.2018.00343
Prediction of Cacao (Theobroma cacao) Resistance to Moniliophthora spp. Diseases via Genome-Wide Association Analysis and Genomic Selection
Abstract
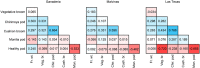
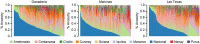
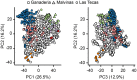
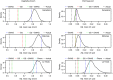
Cacao (Theobroma cacao) is a globally important crop, and its yield is severely restricted by disease. Two of the most damaging diseases, witches' broom disease (WBD) and frosty pod rot disease (FPRD), are caused by a pair of related fungi: Moniliophthora perniciosa and Moniliophthora roreri, respectively. Resistant cultivars are the most effective long-term strategy to address Moniliophthora diseases, but efficiently generating resistant and productive new cultivars will require robust methods for screening germplasm before field testing. Marker-assisted selection (MAS) and genomic selection (GS) provide two potential avenues for predicting the performance of new genotypes, potentially increasing the selection gain per unit time. To test the effectiveness of these two approaches, we performed a genome-wide association study (GWAS) and GS on three related populations of cacao in Ecuador genotyped with a 15K single nucleotide polymorphism (SNP) microarray for three measures of WBD infection (vegetative broom, cushion broom, and chirimoya pod), one of FPRD (monilia pod) and two productivity traits (total fresh weight of pods and % healthy pods produced). GWAS yielded several SNPs associated with disease resistance in each population, but none were significantly correlated with the same trait in other populations. Genomic selection, using one population as a training set to estimate the phenotypes of the remaining two (composed of different families), varied among traits, from a mean prediction accuracy of 0.46 (vegetative broom) to 0.15 (monilia pod), and varied between training populations. Simulations demonstrated that selecting seedlings using GWAS markers alone generates no improvement over selecting at random, but that GS improves the selection process significantly. Our results suggest that the GWAS markers discovered here are not sufficiently predictive across diverse germplasm to be useful for MAS, but that using all markers in a GS framework holds substantial promise in accelerating disease-resistance in cacao.
Keywords: GWAS; SNPs; Theobroma cacao; frosty pod rot; genomic selection; witches’ broom disease.
Figures



References
-
- Aime M. C., Phillips-Mora W. (2005). The causal agents of witches’ broom and frosty pod rot of cacao (chocolate, Theobroma cacao) form a new lineage of Marasmiaceae. Mycologia 97 1012–1022. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

