Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart
- PMID: 29664017
- PMCID: PMC5957472
- DOI: 10.1172/JCI98215
Specialized fibroblast differentiated states underlie scar formation in the infarcted mouse heart
Abstract
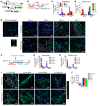
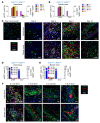
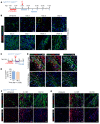
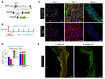
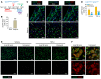
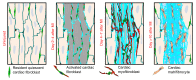
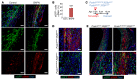
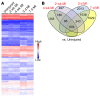
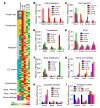
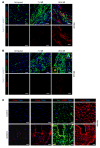
Fibroblasts are a dynamic cell type that achieve selective differentiated states to mediate acute wound healing and long-term tissue remodeling with scarring. With myocardial infarction injury, cardiomyocytes are replaced by secreted extracellular matrix proteins produced by proliferating and differentiating fibroblasts. Here, we employed 3 different mouse lineage-tracing models and stage-specific gene profiling to phenotypically analyze and classify resident cardiac fibroblast dynamics during myocardial infarction injury and stable scar formation. Fibroblasts were activated and highly proliferative, reaching a maximum rate within 2 to 4 days after infarction injury, at which point they expanded 3.5-fold and were maintained long term. By 3 to 7 days, these cells differentiated into myofibroblasts that secreted abundant extracellular matrix proteins and expressed smooth muscle α-actin to structurally support the necrotic area. By 7 to 10 days, myofibroblasts lost proliferative ability and smooth muscle α-actin expression as the collagen-containing extracellular matrix and scar fully matured. However, these same lineage-traced initial fibroblasts persisted within the scar, achieving a new molecular and stable differentiated state referred to as a matrifibrocyte, which was also observed in the scars of human hearts. These cells express common and unique extracellular matrix and tendon genes that are more specialized to support the mature scar.
Keywords: Cardiology; Cardiovascular disease.
Conflict of interest statement
Figures

Comment in
-
A new concept of fibroblast dynamics in post-myocardial infarction remodeling.J Clin Invest. 2018 May 1;128(5):1731-1733. doi: 10.1172/JCI121079. Epub 2018 Apr 16. J Clin Invest. 2018. PMID: 29664015 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

