Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae
- PMID: 29665235
- PMCID: PMC6638102
- DOI: 10.1111/mpp.12692
Genome-wide identification of long non-coding RNAs suggests a potential association with effector gene transcription in Phytophthora sojae
Abstract
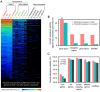
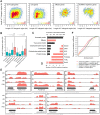
Numerous long non-coding RNAs (lncRNAs) identified and characterized in mammals, plants and fungi have been found to play critical regulatory roles in biological processes. However, little is known about the role of lncRNAs in oomycete plant pathogens, which cause devastating damage to the economy and ecosystems. We used strand-specific RNA sequencing (RNA-seq) to generate a computational pipeline to identify lncRNAs in Phytophthora sojae, a model oomycete plant pathogen. In total, 940 lncRNAs with 1010 isoforms were identified from RNA-seq data obtained from four representative stages of P. sojae. The lncRNAs had shorter transcript lengths, longer exon lengths, fewer numbers of exons, lower GC content and higher minimum free energy values compared with protein-coding genes. lncRNAs in P. sojae exhibited low sequence conservation amongst oomycetes and P. sojae isolates. Transcriptional data indicated that P. sojae lncRNAs tended to be transcribed in a stage-specific manner; representative lncRNAs were validated by semi-quantitative reverse transcription-polymerase chain reaction. Phytophthora sojae lncRNAs were concentrated in gene-sparse regions, and lncRNAs were associated with secreted protein and effector coding genes. The neighbouring genes of lncRNAs encoded various effector family members, and RNA-seq data revealed a correlation between the transcription level of lncRNAs and their neighbouring genes. Our results provide the first comprehensive identification of lncRNAs in oomycetes and suggest a potential association between lncRNAs and effector genes.
Keywords: Phytophthora; RNA-seq; effector; long non-coding RNAs; transcriptional regulation.
© 2018 BSPP and John Wiley & Sons Ltd.
Figures


Similar articles
-
Genome-wide identification of long non-coding RNAs reveals potential association with Phytophthora infestans asexual and sexual development.Microbiol Spectr. 2025 May 6;13(5):e0199824. doi: 10.1128/spectrum.01998-24. Epub 2025 Mar 26. Microbiol Spectr. 2025. PMID: 40135915 Free PMC article.
-
The Pectin Methylesterase Gene Complement of Phytophthora sojae: Structural and Functional Analyses, and the Evolutionary Relationships with Its Oomycete Homologs.PLoS One. 2015 Nov 6;10(11):e0142096. doi: 10.1371/journal.pone.0142096. eCollection 2015. PLoS One. 2015. PMID: 26544849 Free PMC article.
-
Phylogenetic and transcriptional analysis of an expanded bZIP transcription factor family in Phytophthora sojae.BMC Genomics. 2013 Nov 28;14(1):839. doi: 10.1186/1471-2164-14-839. BMC Genomics. 2013. PMID: 24286285 Free PMC article.
-
Long antisense non-coding RNAs and the epigenetic regulation of gene expression.Biomol Concepts. 2013 Aug;4(4):411-5. doi: 10.1515/bmc-2013-0014. Biomol Concepts. 2013. PMID: 25436590 Review.
-
Roles of long noncoding RNAs in brain development, functional diversification and neurodegenerative diseases.Brain Res Bull. 2013 Aug;97:69-80. doi: 10.1016/j.brainresbull.2013.06.001. Epub 2013 Jun 10. Brain Res Bull. 2013. PMID: 23756188 Review.
Cited by
-
Roles of long non-coding RNAs in plant immunity.PLoS Pathog. 2023 May 11;19(5):e1011340. doi: 10.1371/journal.ppat.1011340. eCollection 2023 May. PLoS Pathog. 2023. PMID: 37167200 Free PMC article. Review.
-
Underground communication: Long non-coding RNA signaling in the plant rhizosphere.Plant Commun. 2024 Jul 8;5(7):100927. doi: 10.1016/j.xplc.2024.100927. Epub 2024 Apr 27. Plant Commun. 2024. PMID: 38679911 Free PMC article. Review.
-
The SET domain protein PsKMT3 regulates histone H3K36 trimethylation and modulates effector gene expression in the soybean pathogen Phytophthora sojae.Mol Plant Pathol. 2023 Apr;24(4):346-358. doi: 10.1111/mpp.13301. Epub 2023 Feb 7. Mol Plant Pathol. 2023. PMID: 36748674 Free PMC article.
-
Genome-Wide Identification and Functional Prediction of Long Non-coding RNAs Involved in the Heat Stress Response in Metarhizium robertsii.Front Microbiol. 2019 Oct 9;10:2336. doi: 10.3389/fmicb.2019.02336. eCollection 2019. Front Microbiol. 2019. PMID: 31649657 Free PMC article.
-
Genome-wide identification of long non-coding RNAs reveals potential association with Phytophthora infestans asexual and sexual development.Microbiol Spectr. 2025 May 6;13(5):e0199824. doi: 10.1128/spectrum.01998-24. Epub 2025 Mar 26. Microbiol Spectr. 2025. PMID: 40135915 Free PMC article.
References
-
- Asman, A.K. , Fogelqvist, J. , Vetukuri, R.R. and Dixelius, C. (2016) Phytophthora infestans argonaute 1 binds microRNA and small RNAs from effector genes and transposable elements. New Phytol. 211, 993–1007. - PubMed
-
- Avrova, A.O. , Whisson, S.C. , Pritchard, L. , Venter, E. , De Luca, S. , Hein, I. and Birch, P.R.J. (2007) A novel non‐protein‐coding infection‐specific gene family is clustered throughout the genome of Phytophthora infestans . Microbiology, 153, 747–759. - PubMed
-
- Baldauf, S.L. (2003) The deep roots of eukaryotes. Science, 300, 1703–1706. - PubMed
-
- Baxter, L. , Tripathy, S. , Ishaque, N. , Boot, N. , Cabral, A. , Kemen, E. , Thines, M. , Ah‐Fong, A. , Anderson, R. , Badejoko, W. , Bittner‐Eddy, P. , Boore, J.L. , Chibucos, M.C. , Coates, M. , Dehal, P. , Delehaunty, K. , Dong, S. , Downton, P. , Dumas, B. , Fabro, G. , Fronick, C. , Fuerstenberg, S.I. , Fulton, L. , Gaulin, E. , Govers, F. , Hughes, L. , Humphray, S. , Jiang, R.H.Y. , Judelson, H. , Kamoun, S. , Kyung, K. , Meijer, H. , Minx, P. , Morris, P. , Nelson, J. , Phuntumart, V. , Qutob, D. , Rehmany, A. , Rougon‐Cardoso, A. , Ryden, P. , Torto‐Alalibo, T. , Studholme, D. , Wang, Y. , Win, J. , Wood, J. , Clifton, S.W. , Rogers, J. , Van den Ackerveken, G. , Jones, J.D.G. , McDowell, J.M. , Beynon, J. and Tyler, B.M. (2010) Signatures of adaptation to obligate biotrophy in the Hyaloperonospora arabidopsidis genome. Science, 330, 1549–1551. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

