FBXW7 regulates a mitochondrial transcription program by modulating MITF
- PMID: 29665239
- PMCID: PMC6192859
- DOI: 10.1111/pcmr.12704
FBXW7 regulates a mitochondrial transcription program by modulating MITF
Abstract
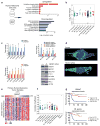
FBXW7 is well characterized as a tumor suppressor in many human cancers including melanoma; however, the mechanisms of tumor-suppressive function have not been fully elucidated. We leveraged two distinct RNA sequencing datasets: human melanoma cell lines (n = 10) with control versus silenced FBXW7 and a cohort of human melanoma tumor samples (n = 51) to define the transcriptomic fingerprint regulated by FBXW7. Here, we report that loss of FBXW7 enhances a mitochondrial gene transcriptional program that is dependent on MITF in human melanoma and confers poor patient outcomes. MITF is a lineage-specific master regulator of melanocytes and together with PGC-1alpha is a marker for melanoma subtypes with dependence for mitochondrial oxidative metabolism. We found that inactivation of FBXW7 elevates MITF protein levels in melanoma cells. In vitro studies examining loss of FBXW7 and MITF alone or in combination showed that FBXW7 is an upstream regulator for the MITF/PGC-1 signaling.
Keywords: FBXW7; MITF; PGC-1alpha; melanoma; metabolism; mitochondria.
© 2018 John Wiley & Sons A/S. Published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

