The ins and outs of vesicular monoamine transporters
- PMID: 29666153
- PMCID: PMC5940252
- DOI: 10.1085/jgp.201711980
The ins and outs of vesicular monoamine transporters
Abstract
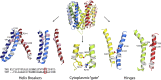
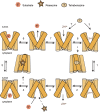
The H+-coupled vesicular monoamine transporter (VMAT) is a transporter essential for life. VMAT mediates packaging of the monoamines serotonin, dopamine, norepinephrine, and histamine from the neuronal cytoplasm into presynaptic vesicles, which is a key step in the regulated release of neurotransmitters. However, a detailed understanding of the mechanism of VMAT function has been limited by the lack of availability of high-resolution structural data. In recent years, a series of studies guided by homology models has revealed significant insights into VMAT function, identifying residues that contribute to the binding site and to specific steps in the transport cycle. Moreover, to characterize the conformational transitions that occur upon binding of the substrate and coupling ion, we have taken advantage of the unique and powerful pharmacology of VMAT as well as of mutants that affect the conformational equilibrium of the protein and shift it toward defined conformations. This has allowed us to identify an important role for the proton gradient in driving a shift from lumen-facing to cytoplasm-facing conformations.
© 2018 Yaffe et al.
Figures

References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

