Biomaterial surface energy-driven ligand assembly strongly regulates stem cell mechanosensitivity and fate on very soft substrates
- PMID: 29666253
- PMCID: PMC5939054
- DOI: 10.1073/pnas.1704543115
Biomaterial surface energy-driven ligand assembly strongly regulates stem cell mechanosensitivity and fate on very soft substrates
Abstract
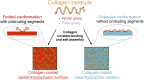
Although mechanisms of cell-material interaction and cellular mechanotransduction are increasingly understood, the mechanical insensitivity of mesenchymal cells to certain soft amorphous biomaterial substrates has remained largely unexplained. We reveal that surface energy-driven supramolecular ligand assembly can regulate mesenchymal stem cell (MSC) sensing of substrate mechanical compliance and subsequent cell fate. Human MSCs were cultured on collagen-coated hydrophobic polydimethylsiloxane (PDMS) and hydrophilic polyethylene-oxide-PDMS (PEO-PDMS) of a range of stiffnesses. Although cell contractility was similarly diminished on soft substrates of both types, cell spreading and osteogenic differentiation occurred only on soft PDMS and not hydrophilic PEO-PDMS (elastic modulus <1 kPa). Substrate surface energy yields distinct ligand topologies with accordingly distinct profiles of recruited transmembrane cell receptors and related focal adhesion signaling. These differences did not differentially regulate Rho-associated kinase activity, but nonetheless regulated both cell spreading and downstream differentiation.
Keywords: PDMS; ligand assembly; mechanobiology; stem cell; surface energy.
Copyright © 2018 the Author(s). Published by PNAS.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Engler AJ, Sen S, Sweeney HL, Discher DE. Matrix elasticity directs stem cell lineage specification. Cell. 2006;126:677–689. - PubMed
-
- Trappmann B, et al. Extracellular-matrix tethering regulates stem-cell fate. Nat Mater. 2012;11:642–649. - PubMed
-
- Prager-Khoutorsky M, et al. Fibroblast polarization is a matrix-rigidity-dependent process controlled by focal adhesion mechanosensing. Nat Cell Biol. 2011;13:1457–1465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

