Chemotactic Cues for NOTCH1-Dependent Leukemia
- PMID: 29666622
- PMCID: PMC5891592
- DOI: 10.3389/fimmu.2018.00633
Chemotactic Cues for NOTCH1-Dependent Leukemia
Abstract
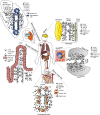
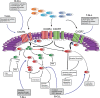
The NOTCH signaling pathway is a conserved signaling cascade that regulates many aspects of development and homeostasis in multiple organ systems. Aberrant activity of this signaling pathway is linked to the initiation and progression of several hematological malignancies, exemplified by T-cell acute lymphoblastic leukemia (T-ALL). Interestingly, frequent non-mutational activation of NOTCH1 signaling has recently been demonstrated in B-cell chronic lymphocytic leukemia (B-CLL), significantly extending the pathogenic significance of this pathway in B-CLL. Leukemia patients often present with high-blood cell counts, diffuse disease with infiltration of the bone marrow, secondary lymphoid organs, and diffusion to the central nervous system (CNS). Chemokines are chemotactic cytokines that regulate migration of cells between tissues and the positioning and interactions of cells within tissue. Homeostatic chemokines and their receptors have been implicated in regulating organ-specific infiltration, but may also directly and indirectly modulate tumor growth. Recently, oncogenic NOTCH1 has been shown to regulate infiltration of leukemic cells into the CNS hijacking the CC-chemokine ligand 19/CC-chemokine receptor 7 chemokine axis. In addition, a crucial role for the homing receptor axis CXC-chemokine ligand 12/CXC-chemokine receptor 4 has been demonstrated in leukemia maintenance and progression. Moreover, the CCL25/CCR9 axis has been implicated in the homing of leukemic cells into the gut, particularly in the presence of phosphatase and tensin homolog tumor suppressor loss. In this review, we summarize the latest developments regarding the role of NOTCH signaling in regulating the chemotactic microenvironmental cues involved in the generation and progression of T-ALL and compare these findings to B-CLL.
Keywords: CXC-chemokine receptor 4; CXCR7; NOTCH; T-cell acute lymphoblastic leukemia; chemokines; infiltration; stromal-derived factor-1.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

