Pre-diagnostic blood immune markers, incidence and progression of B-cell lymphoma and multiple myeloma: Univariate and functionally informed multivariate analyses
- PMID: 29667176
- PMCID: PMC6100111
- DOI: 10.1002/ijc.31536
Pre-diagnostic blood immune markers, incidence and progression of B-cell lymphoma and multiple myeloma: Univariate and functionally informed multivariate analyses
Abstract
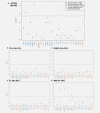
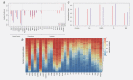
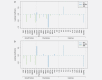
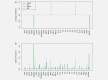
Recent prospective studies have shown that dysregulation of the immune system may precede the development of B-cell lymphomas (BCL) in immunocompetent individuals. However, to date, the studies were restricted to a few immune markers, which were considered separately. Using a nested case-control study within two European prospective cohorts, we measured plasma levels of 28 immune markers in samples collected a median of 6 years before diagnosis (range 2.01-15.97) in 268 incident cases of BCL (including multiple myeloma [MM]) and matched controls. Linear mixed models and partial least square analyses were used to analyze the association between levels of immune marker and the incidence of BCL and its main histological subtypes and to investigate potential biomarkers predictive of the time to diagnosis. Linear mixed model analyses identified associations linking lower levels of fibroblast growth factor-2 (FGF-2 p = 7.2 × 10-4 ) and transforming growth factor alpha (TGF-α, p = 6.5 × 10-5 ) and BCL incidence. Analyses stratified by histological subtypes identified inverse associations for MM subtype including FGF-2 (p = 7.8 × 10-7 ), TGF-α (p = 4.08 × 10-5 ), fractalkine (p = 1.12 × 10-3 ), monocyte chemotactic protein-3 (p = 1.36 × 10-4 ), macrophage inflammatory protein 1-alpha (p = 4.6 × 10-4 ) and vascular endothelial growth factor (p = 4.23 × 10-5 ). Our results also provided marginal support for already reported associations between chemokines and diffuse large BCL (DLBCL) and cytokines and chronic lymphocytic leukemia (CLL). Case-only analyses showed that Granulocyte-macrophage colony stimulating factor levels were consistently higher closer to diagnosis, which provides further evidence of its role in tumor progression. In conclusion, our study suggests a role of growth-factors in the incidence of MM and of chemokine and cytokine regulation in DLBCL and CLL.
Keywords: cytokine; lymphoma; mixed-effect modeling; multiple myeloma; multivariate models; prospective cohort; time to diagnosis.
© 2018 The Authors International Journal of Cancer published by John Wiley & Sons Ltd on behalf of UICC.
Figures
References
-
- Müller AM, Ihorst G, Mertelsmann R, et al. Epidemiology of non‐Hodgkin's lymphoma (NHL): trends, geographic distribution, and etiology. Ann Hematol 2005;84:1–12. - PubMed
-
- Jaffe ES, Harris NL, Stein H, et al. World health organization classification of tumours: pathology and genetics of tumours of hematopoietic and lymphoid tissues. International Agency for Research on Cancer, 2001.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

