Genetic characterization of a novel picorna-like virus in Culex spp. mosquitoes from Mozambique
- PMID: 29669586
- PMCID: PMC5907373
- DOI: 10.1186/s12985-018-0981-z
Genetic characterization of a novel picorna-like virus in Culex spp. mosquitoes from Mozambique
Abstract
Background: Mosquitoes are the potential vectors for a variety of viruses that can cause diseases in the human and animal populations. Viruses in the order Picornavirales infect a broad range of hosts, including mosquitoes. In this study, we aimed to characterize a novel picorna-like virus from the Culex spp. of mosquitoes from the Zambezi Valley of Mozambique.
Methods: The extracted RNA from mosquito pools was pre-amplified with the sequence independent single primer amplification (SISPA) method and subjected to high-throughput sequencing using the Ion Torrent platform. Reads that are classified as Iflaviridae, Picornaviridae and Dicistroviridae were assembled by CodonCode Aligner and SPAdes. Gaps between the viral contigs were sequenced by PCR. The genomic ends were analyzed by 5' and 3' RACE PCRs. The ORF was predicted with the NCBI ORF finder. The conserved domains were identified with ClustalW multiple sequence alignment, and a phylogenetic tree was built with MEGA. The presence of the virus in individual mosquito pools was detected by RT-PCR assay.
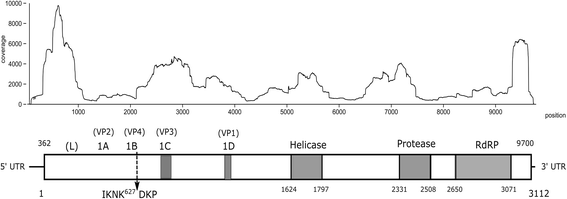
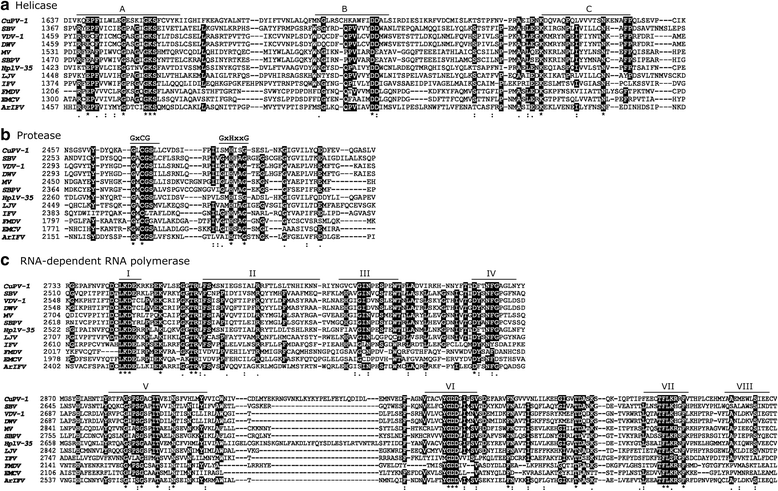
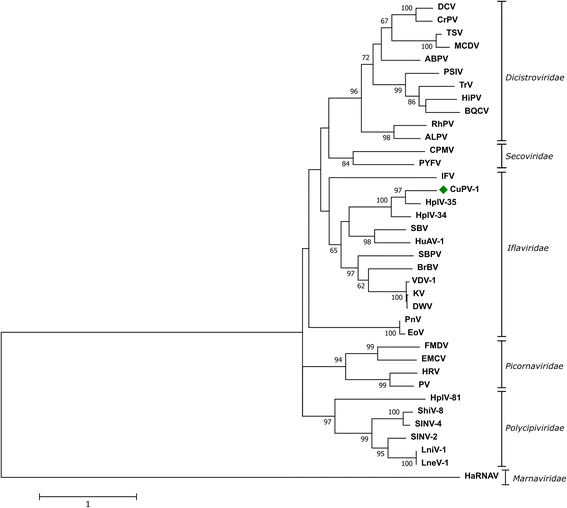
Results: A near full-length viral genome (9740 nt) was obtained in Culex mosquitoes that encoded a complete ORF (3112 aa), named Culex picorna-like virus (CuPV-1). The predicted ORF had 38% similarity to the Hubei picorna-like virus 35. The sequence of the conserved domains, Helicase-Protease-RNA-dependent RNA polymerase, were identified by multiple sequence alignment and found to be at the 3' end, similar to iflaviruses. Phylogenetic analysis of the putative RdRP amino acid sequences indicated that the virus clustered with members of the Iflaviridae family. CuPV-1 was detected in both Culex and Mansonia individual pools with low infection rates.
Conclusions: The study reported a highly divergent, near full-length picorna-like virus genome from Culex spp. mosquitoes from Mozambique. The discovery and characterization of novel viruses in mosquitoes is an initial step, which will provide insights into mosquito-virus interaction mechanisms, genetic diversity and evolution.
Keywords: Culex; Iflavirus; Mosquitoes; Picorna-like virus; RNA virus.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors have declared no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
The complete genome sequence of Perina nuda picorna-like virus, an insect-infecting RNA virus with a genome organization similar to that of the mammalian picornaviruses.Virology. 2002 Mar 15;294(2):312-23. doi: 10.1006/viro.2001.1344. Virology. 2002. PMID: 12009873
-
Detection of Quang Binh virus from mosquitoes in China.Virus Res. 2014 Feb 13;180:31-8. doi: 10.1016/j.virusres.2013.12.005. Epub 2013 Dec 14. Virus Res. 2014. PMID: 24342141
-
Novel strains of Culex flavivirus and Hubei chryso-like virus 1 from the Anopheles mosquito in western Kenya.Virus Res. 2024 Jan 2;339:199266. doi: 10.1016/j.virusres.2023.199266. Epub 2023 Nov 11. Virus Res. 2024. PMID: 37944758 Free PMC article.
-
The Big Bang of picorna-like virus evolution antedates the radiation of eukaryotic supergroups.Nat Rev Microbiol. 2008 Dec;6(12):925-39. doi: 10.1038/nrmicro2030. Epub 2008 Nov 10. Nat Rev Microbiol. 2008. PMID: 18997823 Review.
-
The discovery and global distribution of novel mosquito-associated viruses in the last decade (2007-2017).Rev Med Virol. 2019 Nov;29(6):e2079. doi: 10.1002/rmv.2079. Epub 2019 Aug 13. Rev Med Virol. 2019. PMID: 31410931 Review.
Cited by
-
Metagenomic sequencing reveals viral abundance and diversity in mosquitoes from the Shaanxi-Gansu-Ningxia region, China.PLoS Negl Trop Dis. 2021 Apr 26;15(4):e0009381. doi: 10.1371/journal.pntd.0009381. eCollection 2021 Apr. PLoS Negl Trop Dis. 2021. PMID: 33901182 Free PMC article.
-
Deciphering the Virome of Culex vishnui Subgroup Mosquitoes, the Major Vectors of Japanese Encephalitis, in Japan.Viruses. 2020 Feb 28;12(3):264. doi: 10.3390/v12030264. Viruses. 2020. PMID: 32121094 Free PMC article.
-
Viromics on Honey-Baited FTA Cards as a New Tool for the Detection of Circulating Viruses in Mosquitoes.Viruses. 2020 Feb 29;12(3):274. doi: 10.3390/v12030274. Viruses. 2020. PMID: 32121402 Free PMC article.
-
Shotgun Metagenomic Sequencing Reveals Virome Composition of Mosquitoes from a Transition Ecosystem of North-Northeast Brazil.Genes (Basel). 2023 Jul 14;14(7):1443. doi: 10.3390/genes14071443. Genes (Basel). 2023. PMID: 37510347 Free PMC article.
-
An Exploration of the Viral Coverage of Mosquito Viromes Using Meta-Viromic Sequencing: A Systematic Review and Meta-Analysis.Microorganisms. 2024 Sep 14;12(9):1899. doi: 10.3390/microorganisms12091899. Microorganisms. 2024. PMID: 39338573 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

