Two distinct sites of client protein interaction with the chaperone cpSRP43
- PMID: 29669809
- PMCID: PMC5995501
- DOI: 10.1074/jbc.RA118.002215
Two distinct sites of client protein interaction with the chaperone cpSRP43
Abstract
Integral membrane proteins are prone to aggregation and misfolding in aqueous environments and therefore require binding by molecular chaperones during their biogenesis. Chloroplast signal recognition particle 43 (cpSRP43) is an ATP-independent chaperone required for the biogenesis of the most abundant class of membrane proteins, the light-harvesting chlorophyll a/b-binding proteins (LHCPs). Previous work has shown that cpSRP43 specifically recognizes an L18 loop sequence conserved among LHCP paralogs. However, how cpSRP43 protects the transmembrane domains (TMDs) of LHCP from aggregation was unclear. In this work, alkylation-protection and site-specific cross-linking experiments found that cpSRP43 makes extensive contacts with all the TMDs in LHCP. Site-directed mutagenesis identified a class of cpSRP43 mutants that bind tightly to the L18 sequence but are defective in chaperoning full-length LHCP. These mutations mapped to hydrophobic surfaces on or near the bridging helix and the β-hairpins lining the ankyrin repeat motifs of cpSRP43, suggesting that these regions are potential sites for interaction with the client TMDs. Our results suggest a working model for client protein interactions in this membrane protein chaperone.
Keywords: chaperone; chloroplast; membrane protein; protein targeting; signal recognition particle (SRP).
© 2018 McAvoy et al.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
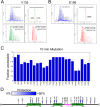





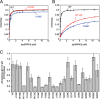

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

