Molecular Preadaptation to Antimony Resistance in Leishmania donovani on the Indian Subcontinent
- PMID: 29669889
- PMCID: PMC5907651
- DOI: 10.1128/mSphere.00548-17
Molecular Preadaptation to Antimony Resistance in Leishmania donovani on the Indian Subcontinent
Abstract
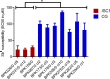
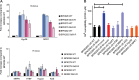
Antimonials (Sb) were used for decades for chemotherapy of visceral leishmaniasis (VL). Now abandoned in the Indian subcontinent (ISC) because of Leishmania donovani resistance, this drug offers a unique model for understanding drug resistance dynamics. In a previous phylogenomic study, we found two distinct populations of L. donovani: the core group (CG) in the Gangetic plains and ISC1 in the Nepalese highlands. Sb resistance was only encountered within the CG, and a series of potential markers were identified. Here, we analyzed the development of resistance to trivalent antimonials (SbIII) upon experimental selection in ISC1 and CG strains. We observed that (i) baseline SbIII susceptibility of parasites was higher in ISC1 than in the CG, (ii) time to SbIII resistance was higher for ISC1 parasites than for CG strains, and (iii) untargeted genomic and metabolomic analyses revealed molecular changes along the selection process: these were more numerous in ISC1 than in the CG. Altogether these observations led to the hypothesis that CG parasites are preadapted to SbIII resistance. This hypothesis was experimentally confirmed by showing that only wild-type CG strains could survive a direct exposure to the maximal concentration of SbIII The main driver of this preadaptation was shown to be MRPA, a gene involved in SbIII sequestration and amplified in an intrachromosomal amplicon in all CG strains characterized so far. This amplicon emerged around 1850 in the CG, well before the implementation of antimonials for VL chemotherapy, and we discuss here several hypotheses of selective pressure that could have accompanied its emergence.IMPORTANCE The "antibiotic resistance crisis" is a major challenge for scientists and medical professionals. This steady rise in drug-resistant pathogens also extends to parasitic diseases, with antimony being the first anti-Leishmania drug that fell in the Indian subcontinent (ISC). Leishmaniasis is a major but neglected infectious disease with limited therapeutic options. Therefore, understanding how parasites became resistant to antimonials is of commanding importance. In this study, we experimentally characterized the dynamics of this resistance acquisition and show for the first time that some Leishmania populations of the ISC were preadapted to antimony resistance, likely driven by environmental factors or by drugs used in the 19th century.
Keywords: Leishmania; antimonials; drug resistance mechanisms; genomics; metabolomics.
Copyright © 2018 Dumetz et al.
Figures




References
-
- Mookerjee Basu J, Mookerjee A, Sen P, Bhaumik S, Sen P, Banerjee S, Naskar K, Choudhuri SK, Saha B, Raha S, Roy S. 2006. Sodium-antimony gluconate induces generation of reactive oxygen species and nitric oxide via phosphoinositide 3-kinase and mitogen-activated protein kinase activation in Leishmania donovani-infected macrophages. Antimicrob Agents Chemother 50:1788–1797. doi:10.1128/AAC.50.5.1788-1797.2006. - DOI - PMC - PubMed
-
- Mandal G, Wyllie S, Singh N, Sundar S, Fairlamb AH, Chatterjee M. 2007. Increased levels of thiols protect antimony unresponsive Leishmania donovani field isolates against reactive oxygen species generated by trivalent antimony. Parasitology 134:1679–1687. doi:10.1017/S0031182007003150. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
