Generation and testing of clinical-grade exosomes for pancreatic cancer
- PMID: 29669940
- PMCID: PMC5931131
- DOI: 10.1172/jci.insight.99263
Generation and testing of clinical-grade exosomes for pancreatic cancer
Abstract
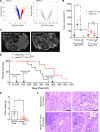
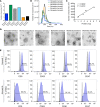
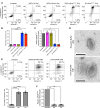
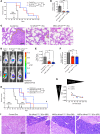
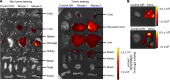
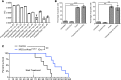
Exosomes are extracellular vesicles produced by all cells with a remarkable ability to efficiently transfer genetic material, including exogenously loaded siRNA, to cancer cells. Here, we report on a bioreactor-based, large-scale production of clinical-grade exosomes employing good manufacturing practice (GMP) standards. A standard operating procedure was established to generate engineered exosomes with the ability to target oncogenic Kras (iExosomes). The clinical-grade GMP iExosomes were tested in multiple in vitro and in vivo studies to confirm suppression of oncogenic Kras and an increase in the survival of several mouse models with pancreatic cancer. We perform studies to determine the shelf life, biodistribution, toxicology profile, and efficacy in combination with chemotherapy to inform future clinical testing of GMP iExosomes. Collectively, this report illustrates the process and feasibility of generating clinical-grade exosomes for various therapies of human diseases.
Keywords: Cancer gene therapy; Oncology.
Conflict of interest statement
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

