Micro-scale intermixing: a requisite for stable and synergistic co-establishment in a four-species biofilm
- PMID: 29670216
- PMCID: PMC6052071
- DOI: 10.1038/s41396-018-0112-2
Micro-scale intermixing: a requisite for stable and synergistic co-establishment in a four-species biofilm
Abstract
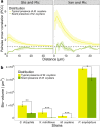
Microorganisms frequently coexist in complex multispecies communities, where they distribute non-randomly, reflective of the social interactions that occur. It is therefore important to understand how social interactions and local spatial organization influences multispecies biofilm succession. Here the localization of species pairs was analyzed in three dimensions in a reproducible four-species biofilm model, to study the impact of spatial positioning of individual species on the temporal development of the community. We found, that as the biofilms developed, species pairs exhibited distinct intermixing patterns unique to the four-member biofilms. Higher biomass and more intermixing were found in four-species biofilms compared to biofilms with fewer species. Intriguingly, in local regions within the four member biofilms where Microbacterium oxydans was scant, both biomass and intermixing of all species were lowered, compared to regions where M. oxydans was present at typical densities. Our data suggest that Xanthomonas retroflexus and M. oxydans, both low abundant biofilm-members, intermixed continuously during the development of the four-species biofilm, hereby facilitating their own establishment. In turn, this seems to have promoted distinct spatial organization of Stenotrophomonas rhizophila and Paenibacillus amylolyticus enabling enhanced growth of all four species. Here local intermixing of bacteria advanced the temporal development of a multi-species biofilm.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

