ANLN functions as a key candidate gene in cervical cancer as determined by integrated bioinformatic analysis
- PMID: 29670400
- PMCID: PMC5896649
- DOI: 10.2147/CMAR.S162813
ANLN functions as a key candidate gene in cervical cancer as determined by integrated bioinformatic analysis
Abstract
Background: Cervical cancer, one of the leading causes of female deaths, remains a top cause of mortality in gynecologic oncology and tends to affect younger individuals. However, the pathogenesis of cervical cancer is still far from clear. Given the high incidence and mortality of cervical cancer, uncovering the causes and pathogenesis as well as identifying novel biomarkers are of great significance and are desperately needed.
Materials and methods: First, raw data were downloaded from the Gene Expression Omnibus database. The Robuse Multi-Array Average algorithm and combat function of the sva package were subsequently applied to preprocess and remove batch effects. Differentially expressed genes (DEGs) analyzed with the limma package were followed by gene ontology and pathway analysis, and a protein-protein interaction (PPI) network based on the STRING website and the Cytoscape software was constructed. Weighted Correlation Network Analysis (WGCNA) was utilized to build the coexpression network. Subsequently, UALCAN websites were employed to conduct survival analysis. Finally, the oncomine database was used to validate the expression of ANLN in other datasets.
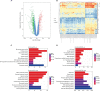
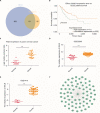
Results: GSE29570 and GSE89657, including 49 cervical cancer tissues and 20 normal cervical tissues, were screened as the datasets. Three-hundred-twenty-four DEGs were identified and, among them, 123 were upregulated, while 201 were downregulated. The DEGs PPI network complex, contained 305 nodes and 4,962 edges, and 8 clusters were calculated according to k-core =2. Among them, cluster 1, which had 65 nodes and 1,780 edges, had the highest score in these clusters. In coexpression analysis, there were 86 hubgenes from the Brown modules that were chosen for further analysis. Sixty-one key genes were identified as the intersecting genes of the Brown module of WGCNA and DEGs. In survival analysis, only ANLN was a prognostic factor, and the survival was significantly better in the low-expression ANLN group.
Conclusion: Our study suggested that ANLN may be a potential tumor oncogene and could serve as a biomarker for predicting the prognosis of cervical cancer patients.
Keywords: ANLN; WGCNA; bioinformatics analysis; cervical cancer.
Conflict of interest statement
Disclosure The authors report no conflicts of interest in this work.
Figures


References
-
- Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLO- BOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. - PubMed
-
- Brinton LA, Herrero R, Reeves WC, de Britton RC, Gaitan E, Tenorio F. Risk factors for cervical cancer by histology. Gynecol Oncol. 1993;51(3):301–306. - PubMed
-
- Jeong H, Mason SP, Barabási AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411(6833):41–42. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

