Data Mining and Computational Modeling of High-Throughput Screening Datasets
- PMID: 29671272
- PMCID: PMC6181121
- DOI: 10.1007/978-1-4939-7724-6_14
Data Mining and Computational Modeling of High-Throughput Screening Datasets
Abstract
We are now seeing the benefit of investments made over the last decade in high-throughput screening (HTS) that is resulting in large structure activity datasets entering public and open databases such as ChEMBL and PubChem. The growth of academic HTS screening centers and the increasing move to academia for early stage drug discovery suggests a great need for the informatics tools and methods to mine such data and learn from it. Collaborative Drug Discovery, Inc. (CDD) has developed a number of tools for storing, mining, securely and selectively sharing, as well as learning from such HTS data. We present a new web based data mining and visualization module directly within the CDD Vault platform for high-throughput drug discovery data that makes use of a novel technology stack following modern reactive design principles. We also describe CDD Models within the CDD Vault platform that enables researchers to share models, share predictions from models, and create models from distributed, heterogeneous data. Our system is built on top of the Collaborative Drug Discovery Vault Activity and Registration data repository ecosystem which allows users to manipulate and visualize thousands of molecules in real time. This can be performed in any browser on any platform. In this chapter we present examples of its use with public datasets in CDD Vault. Such approaches can complement other cheminformatics tools, whether open source or commercial, in providing approaches for data mining and modeling of HTS data.
Keywords: ADME; Bayesian models; CDD models; CDD vault; Collaborative database; Data mining; Visualization.
Figures



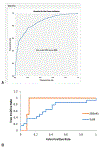
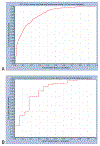
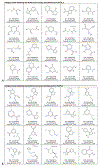
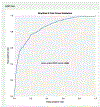
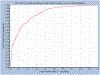
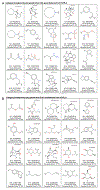

References
-
- Macarron R; Banks MN; Bojanic D; Burns DJ; Cirovic DA; Garyantes T; Green DV; Hertzberg RP; Janzen WP; Paslay JW; Schopfer U; Sittampalam GS , Impact of High-Throughput Screening in Biomedical Research. Nat Rev Drug Discov 2011, 10, 188–195. - PubMed
-
- Ekins S; Waller CL; Bradley MP; Clark AM; Williams AJ, Four Disruptive Strategies for Removing Drug Discovery Bottlenecks Drug Disc Today 2013, 18, 265–271. - PubMed
-
- Kaiser J, National Institutes of Health. Drug-Screening Program Looking for a Home. Science 2011, 334, 299. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

