Foldamer Tertiary Structure through Sequence-Guided Protein Backbone Alteration
- PMID: 29672021
- PMCID: PMC5953838
- DOI: 10.1021/acs.accounts.8b00048
Foldamer Tertiary Structure through Sequence-Guided Protein Backbone Alteration
Abstract
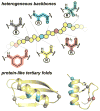
The prospect of recreating the complex structural hierarchy of protein folding in synthetic oligomers with backbones that are artificial in covalent structure ("foldamers") has long fascinated chemists. Foldamers offer complex functions from biostable scaffolds and have found widespread applications in fields from biomedical to materials science. Most precedent has focused on isolated secondary structures or their assemblies. In considering the goal of complex protein-like tertiary folding patterns, a key barrier became apparent. How does one design a backbone with covalent connectivity and a sequence of side-chain functional groups that will support defined intramolecular packing of multiple artificial secondary structures? Two developments were key to overcoming this challenge. First was the recognition of the power of blending α-amino acid residues with monomers differing in backbone connectivity to create "heterogeneous-backbone" foldamers. Second was the finding that replacing some of the natural α-residues in a biological sequence with artificial-backbone variants can result in a mimic that retains both the fold and function of the native sequence and, in some cases, gains advantageous characteristics. Taken together, these precedents lead to a view of a protein as chemical entity having two orthogonal sequences: a sequence of side-chain functional groups and a separate sequence of backbone units displaying those functional groups. In this Account, we describe our lab's work over the last ∼10 years to leverage the above concept of protein sequence duality in order to develop design principles for constructing heterogeneous-backbone foldamers that adopt complex protein-like tertiary folds. Fundamental to the approach is the utilization of a variety of artificial building blocks (e.g., d-α-residues, Cα-Me-α-residues, N-Me-α-residues, β-residues, γ-residues, δ-residues, polymer segments) in concert, replacing a fraction of α-residues in a given prototype sequence. We provide an overview of the state-of-the-art in terms of design principles for choosing substitutions based on consideration of local secondary structure and retention of key side-chain functional groups. We survey high-resolution structures of backbone-modified proteins to illustrate how diverse artificial moieties are accommodated in tertiary fold contexts. We detail efforts to elucidate how backbone alteration impacts folding thermodynamics and describe how such data informs the development of improved design rules. Collectively, a growing body of results by our lab and others spanning multiple protein systems suggests there is a great deal of plasticity with respect to the backbone chemical structures upon which sequence-encoded tertiary folds can manifest. Moreover, these efforts suggest sequence-guided backbone alteration as a broadly applicable strategy for generating foldamers with complex tertiary folding patterns. We conclude by offering some perspective regarding the near future of this field, in terms of unanswered questions, technological needs, and opportunities for new areas of inquiry.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
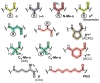


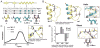
Similar articles
-
Foldamers with heterogeneous backbones.Acc Chem Res. 2008 Oct;41(10):1399-408. doi: 10.1021/ar800009n. Epub 2008 Jul 1. Acc Chem Res. 2008. PMID: 18590282 Free PMC article.
-
Heterogeneous-Backbone Foldamer Mimics of Zinc Finger Tertiary Structure.J Am Chem Soc. 2017 Jun 14;139(23):7931-7938. doi: 10.1021/jacs.7b03114. Epub 2017 Jun 5. J Am Chem Soc. 2017. PMID: 28509549 Free PMC article.
-
Heterogeneous-Backbone Foldamer Mimics of a Computationally Designed, Disulfide-Rich Miniprotein.Chembiochem. 2019 Jan 2;20(1):103-110. doi: 10.1002/cbic.201800558. Epub 2018 Nov 27. Chembiochem. 2019. PMID: 30326175 Free PMC article.
-
Foldectures: 3D Molecular Architectures from Self-Assembly of Peptide Foldamers.Acc Chem Res. 2017 Apr 18;50(4):832-841. doi: 10.1021/acs.accounts.6b00545. Epub 2017 Feb 13. Acc Chem Res. 2017. PMID: 28191927 Review.
-
Protein backbone engineering as a strategy to advance foldamers toward the frontier of protein-like tertiary structure.Org Biomol Chem. 2014 Nov 28;12(44):8796-802. doi: 10.1039/c4ob01769b. Org Biomol Chem. 2014. PMID: 25285575 Free PMC article. Review.
Cited by
-
Polymers for Disrupting Protein-Protein Interactions: Where Are We and Where Should We Be?Biomacromolecules. 2024 Oct 14;25(10):6229-6249. doi: 10.1021/acs.biomac.4c00850. Epub 2024 Sep 10. Biomacromolecules. 2024. PMID: 39254158 Review.
-
Folding-controlled assembly of ortho-phenylene-based macrocycles.Chem Sci. 2021 Apr 14;12(20):6992-7002. doi: 10.1039/d1sc01270c. Chem Sci. 2021. PMID: 34123327 Free PMC article.
-
Chemical Shifts of Artificial Monomers Used to Construct Heterogeneous-Backbone Protein Mimetics in Random Coil and Folded States.Pept Sci (Hoboken). 2023 Mar;115(2):e24297. doi: 10.1002/pep2.24297. Epub 2022 Nov 12. Pept Sci (Hoboken). 2023. PMID: 37397503 Free PMC article.
-
α-Methylation Enables the X-ray Crystallographic Observation of Oligomeric Assemblies Formed by a β-Hairpin Peptide Derived from Aβ.J Org Chem. 2025 Jan 10;90(1):394-400. doi: 10.1021/acs.joc.4c02344. Epub 2024 Dec 17. J Org Chem. 2025. PMID: 39689228 Free PMC article.
-
Structural and Functional Mimicry of the Antimicrobial Defensin Plectasin by Analogues with Engineered Backbone Composition.Chembiochem. 2025 Feb 3;26(5):e202400951. doi: 10.1002/cbic.202400951. Epub 2025 Jan 9. Chembiochem. 2025. PMID: 39714882 Free PMC article.
References
-
- Gellman SH. Foldamers: A Manifesto. Acc Chem Res. 1998;31:173–180.
-
- Guichard G, Huc I. Synthetic Foldamers. Chem Commun. 2011;47:5933–5941. - PubMed
-
- Martinek TA, Fulop F. Peptidic Foldamers: Ramping Up Diversity. Chem Soc Rev. 2012;41:687–702. - PubMed
-
- Gopalakrishnan R, Frolov AI, Knerr L, Drury WJ, Valeur E. Therapeutic Potential of Foldamers: From Chemical Biology Tools To Drug Candidates? J Med Chem. 2016;59:9599–9621. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

