Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana
- PMID: 29672610
- PMCID: PMC5929574
- DOI: 10.1371/journal.pgen.1007337
Threshold-dependent repression of SPL gene expression by miR156/miR157 controls vegetative phase change in Arabidopsis thaliana
Abstract
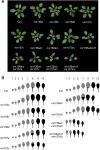
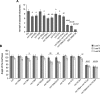
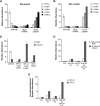
Vegetative phase change is regulated by a decrease in the abundance of the miRNAs, miR156 and miR157, and the resulting increase in the expression of their targets, SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) transcription factors. To determine how miR156/miR157 specify the quantitative and qualitative changes in leaf morphology that occur during vegetative phase change, we measured their abundance in successive leaves and characterized the phenotype of mutations in different MIR156 and MIR157 genes. miR156/miR157 decline rapidly between leaf 1&2 and leaf 3 and decrease more slowly after this point. The amount of miR156/miR157 in leaves 1&2 greatly exceeds the threshold required to specify their identity. Subsequent leaves have relatively low levels of miR156/miR157 and are sensitive to small changes in their abundance. In these later-formed leaves, the amount of miR156/miR157 is close to the threshold required to specify juvenile vs. adult identity; a relatively small decrease in the abundance of miR156/157 in these leaves produces a disproportionately large increase in SPL proteins and a significant change in leaf morphology. miR157 is more abundant than miR156 but has a smaller effect on shoot morphology and SPL gene expression than miR156. This may be attributable to the inefficiency with which miR157 is loaded onto AGO1, as well as to the presence of an extra nucleotide at the 5' end of miR157 that is mis-paired in the miR157:SPL13 duplex. miR156 represses different targets by different mechanisms: it regulates SPL9 by a combination of transcript cleavage and translational repression and regulates SPL13 primarily by translational repression. Our results offer a molecular explanation for the changes in leaf morphology that occur during shoot development in Arabidopsis and provide new insights into the mechanism by which miR156 and miR157 regulate gene expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures








References
-
- Telfer A, Bollman KM, Poethig RS (1997) Phase change and the regulation of trichome distribution in Arabidopsis thaliana. Development 124: 645–654. - PubMed
-
- Willmann MR, Poethig RS (2011) The effect of the floral repressor FLC on the timing and progression of vegetative phase change in Arabidopsis. Development 138: 677–685. doi: 10.1242/dev.057448 - DOI - PMC - PubMed
-
- Wu G, Poethig RS (2006) Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 133: 3539–3547. doi: 10.1242/dev.02521 - DOI - PMC - PubMed
-
- Usami T, Horiguchi G, Yano S, Tsukaya H (2009) The more and smaller cells mutants of Arabidopsis thaliana identify novel roles for SQUAMOSA PROMOTER BINDING PROTEIN-LIKE genes in the control of heteroblasty. Development 136: 955–964. doi: 10.1242/dev.028613 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

