Identification of residue pairing in interacting β-strands from a predicted residue contact map
- PMID: 29673311
- PMCID: PMC5907701
- DOI: 10.1186/s12859-018-2150-1
Identification of residue pairing in interacting β-strands from a predicted residue contact map
Abstract
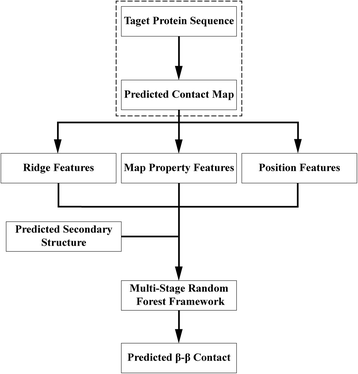
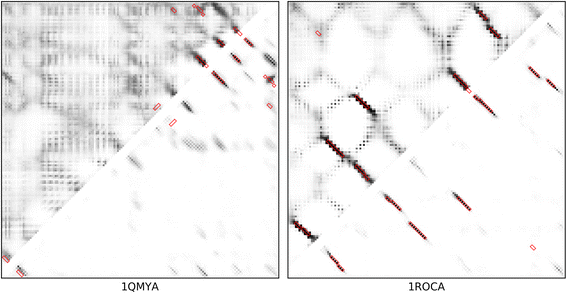
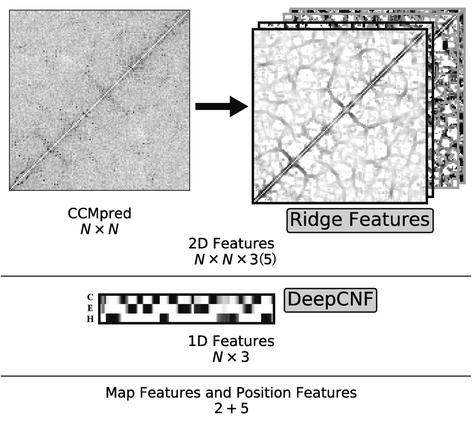
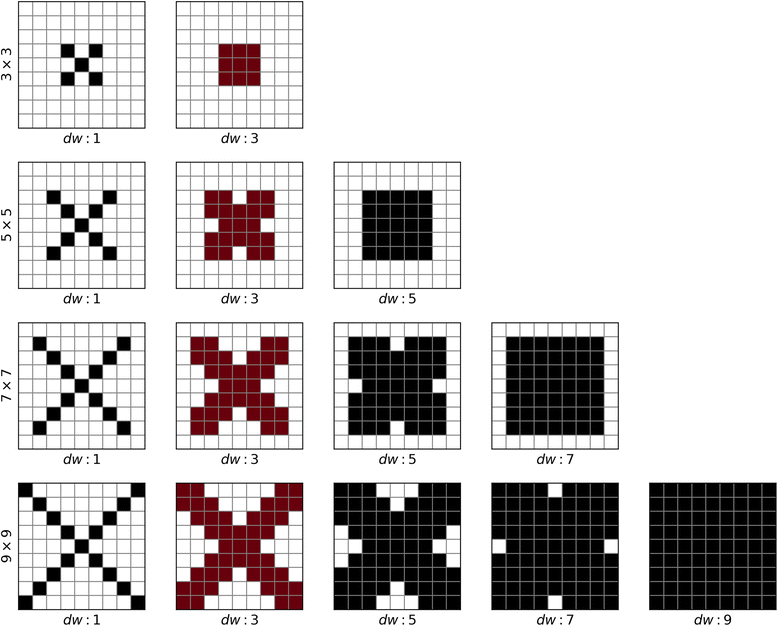
Background: Despite the rapid progress of protein residue contact prediction, predicted residue contact maps frequently contain many errors. However, information of residue pairing in β strands could be extracted from a noisy contact map, due to the presence of characteristic contact patterns in β-β interactions. This information may benefit the tertiary structure prediction of mainly β proteins. In this work, we propose a novel ridge-detection-based β-β contact predictor to identify residue pairing in β strands from any predicted residue contact map.
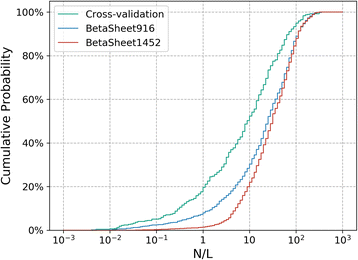
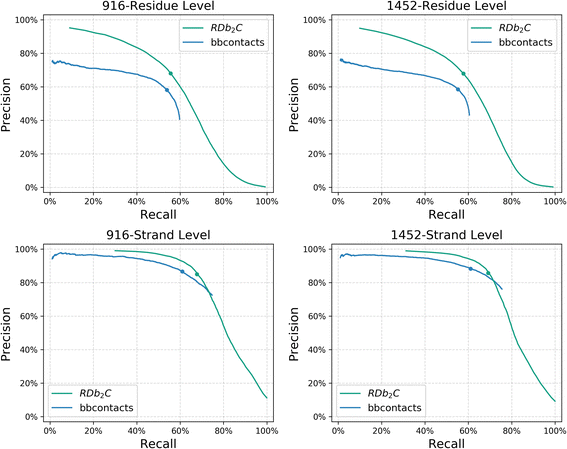
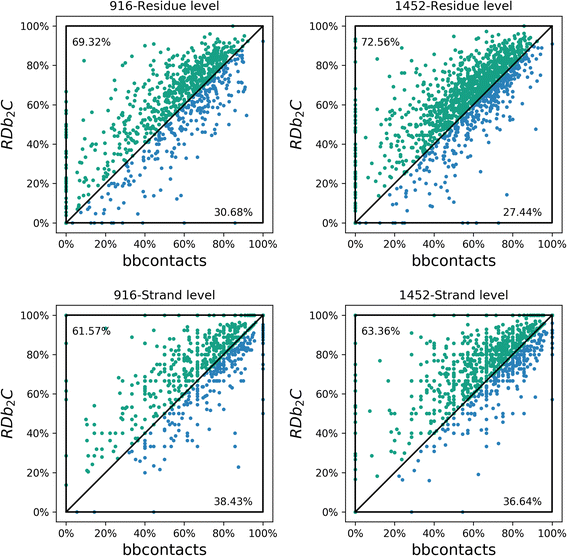
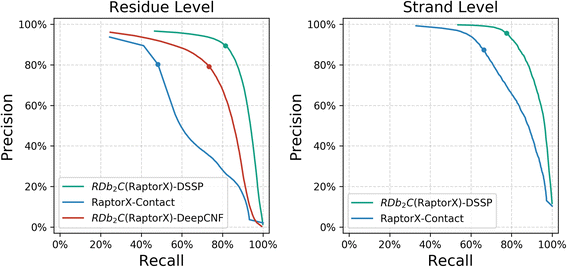
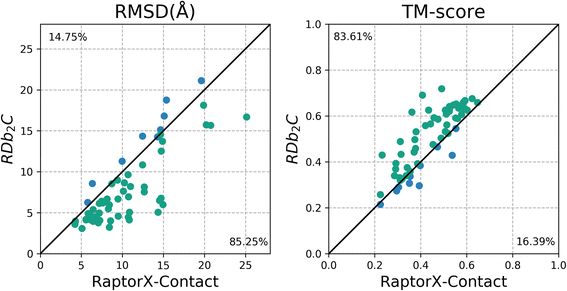
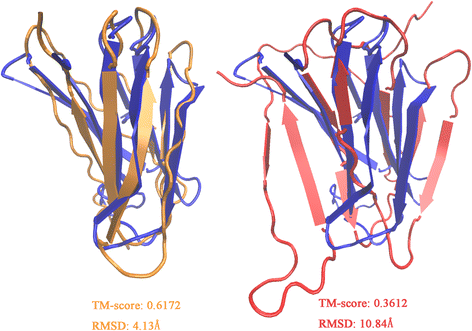
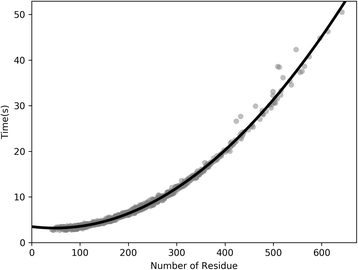
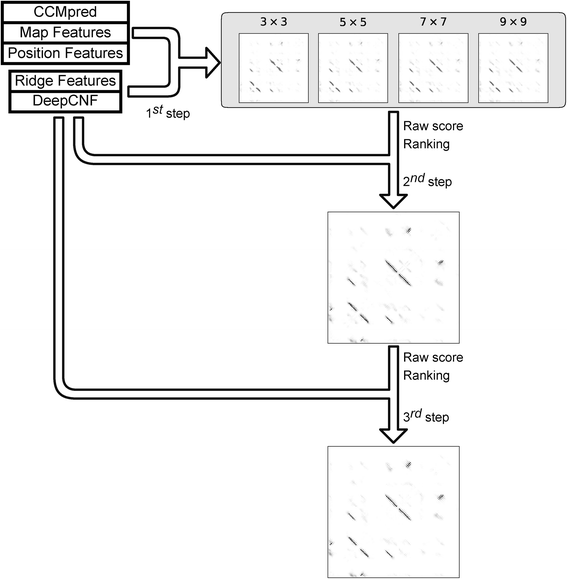
Results: Our algorithm RDb2C adopts ridge detection, a well-developed technique in computer image processing, to capture consecutive residue contacts, and then utilizes a novel multi-stage random forest framework to integrate the ridge information and additional features for prediction. Starting from the predicted contact map of CCMpred, RDb2C remarkably outperforms all state-of-the-art methods on two conventional test sets of β proteins (BetaSheet916 and BetaSheet1452), and achieves F1-scores of ~ 62% and ~ 76% at the residue level and strand level, respectively. Taking the prediction of the more advanced RaptorX-Contact as input, RDb2C achieves impressively higher performance, with F1-scores reaching ~ 76% and ~ 86% at the residue level and strand level, respectively. In a test of structural modeling using the top 1 L predicted contacts as constraints, for 61 mainly β proteins, the average TM-score achieves 0.442 when using the raw RaptorX-Contact prediction, but increases to 0.506 when using the improved prediction by RDb2C.
Conclusion: Our method can significantly improve the prediction of β-β contacts from any predicted residue contact maps. Prediction results of our algorithm could be directly applied to effectively facilitate the practical structure prediction of mainly β proteins.
Availability: All source data and codes are available at http://166.111.152.91/Downloads.html or the GitHub address of https://github.com/wzmao/RDb2C .
Keywords: Contact map; Protein structure prediction; Random forest; Residue contact prediction; Ridge detection; β-β pairing.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Figures

Similar articles
-
RDb2C2: an improved method to identify the residue-residue pairing in β strands.BMC Bioinformatics. 2020 Apr 3;21(1):133. doi: 10.1186/s12859-020-3476-z. BMC Bioinformatics. 2020. PMID: 32245403 Free PMC article.
-
Efficient dynamic programming algorithm with prior knowledge for protein β-strand alignment.J Theor Biol. 2017 Mar 21;417:43-50. doi: 10.1016/j.jtbi.2017.01.018. Epub 2017 Jan 18. J Theor Biol. 2017. PMID: 28108305
-
Accurate De Novo Prediction of Protein Contact Map by Ultra-Deep Learning Model.PLoS Comput Biol. 2017 Jan 5;13(1):e1005324. doi: 10.1371/journal.pcbi.1005324. eCollection 2017 Jan. PLoS Comput Biol. 2017. PMID: 28056090 Free PMC article.
-
Prediction of Structures and Interactions from Genome Information.Adv Exp Med Biol. 2018;1105:123-152. doi: 10.1007/978-981-13-2200-6_9. Adv Exp Med Biol. 2018. PMID: 30617827 Review.
-
The trRosetta server for fast and accurate protein structure prediction.Nat Protoc. 2021 Dec;16(12):5634-5651. doi: 10.1038/s41596-021-00628-9. Epub 2021 Nov 10. Nat Protoc. 2021. PMID: 34759384 Review.
Cited by
-
RDb2C2: an improved method to identify the residue-residue pairing in β strands.BMC Bioinformatics. 2020 Apr 3;21(1):133. doi: 10.1186/s12859-020-3476-z. BMC Bioinformatics. 2020. PMID: 32245403 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

