Multiple large-scale gene and genome duplications during the evolution of hexapods
- PMID: 29674453
- PMCID: PMC5939055
- DOI: 10.1073/pnas.1710791115
Multiple large-scale gene and genome duplications during the evolution of hexapods
Abstract
Polyploidy or whole genome duplication (WGD) is a major contributor to genome evolution and diversity. Although polyploidy is recognized as an important component of plant evolution, it is generally considered to play a relatively minor role in animal evolution. Ancient polyploidy is found in the ancestry of some animals, especially fishes, but there is little evidence for ancient WGDs in other metazoan lineages. Here we use recently published transcriptomes and genomes from more than 150 species across the insect phylogeny to investigate whether ancient WGDs occurred during the evolution of Hexapoda, the most diverse clade of animals. Using gene age distributions and phylogenomics, we found evidence for 18 ancient WGDs and six other large-scale bursts of gene duplication during insect evolution. These bursts of gene duplication occurred in the history of lineages such as the Lepidoptera, Trichoptera, and Odonata. To further corroborate the nature of these duplications, we evaluated the pattern of gene retention from putative WGDs observed in the gene age distributions. We found a relatively strong signal of convergent gene retention across many of the putative insect WGDs. Considering the phylogenetic breadth and depth of the insect phylogeny, this observation is consistent with polyploidy as we expect dosage balance to drive the parallel retention of genes. Together with recent research on plant evolution, our hexapod results suggest that genome duplications contributed to the evolution of two of the most diverse lineages of eukaryotes on Earth.
Keywords: genome duplication; genomics; hexapods; insects; polyploidy.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
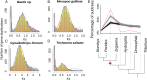
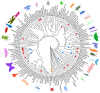
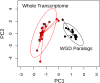
Comment in
-
Reply to Nakatani and McLysaght: Analyzing deep duplication events.Proc Natl Acad Sci U S A. 2019 Feb 5;116(6):1819-1820. doi: 10.1073/pnas.1819227116. Epub 2019 Jan 23. Proc Natl Acad Sci U S A. 2019. PMID: 30674658 Free PMC article. No abstract available.
References
-
- Barker MS, Husband BC, Pires JC. Spreading Winge and flying high: The evolutionary importance of polyploidy after a century of study. Am J Bot. 2016;103:1139–1145. - PubMed
-
- Van de Peer Y, Mizrachi E, Marchal K. The evolutionary significance of polyploidy. Nat Rev Genet. 2017;18:411–424. - PubMed
-
- Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

