Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq
- PMID: 29674595
- PMCID: PMC5949869
- DOI: 10.1126/science.aao4750
Developmental and oncogenic programs in H3K27M gliomas dissected by single-cell RNA-seq
Abstract
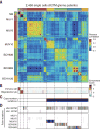
Gliomas with histone H3 lysine27-to-methionine mutations (H3K27M-glioma) arise primarily in the midline of the central nervous system of young children, suggesting a cooperation between genetics and cellular context in tumorigenesis. Although the genetics of H3K27M-glioma are well characterized, their cellular architecture remains uncharted. We performed single-cell RNA sequencing in 3321 cells from six primary H3K27M-glioma and matched models. We found that H3K27M-glioma primarily contain cells that resemble oligodendrocyte precursor cells (OPC-like), whereas more differentiated malignant cells are a minority. OPC-like cells exhibit greater proliferation and tumor-propagating potential than their more differentiated counterparts and are at least in part sustained by PDGFRA signaling. Our study characterizes oncogenic and developmental programs in H3K27M-glioma at single-cell resolution and across genetic subclones, suggesting potential therapeutic targets in this disease.
Copyright © 2018 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Conflict of interest statement
Figures
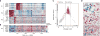


Comment in
-
Inverted architecture.Nat Rev Cancer. 2018 Jul;18(7):405. doi: 10.1038/s41568-018-0025-4. Nat Rev Cancer. 2018. PMID: 29802352 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

