Culturing of female bladder bacteria reveals an interconnected urogenital microbiota
- PMID: 29674608
- PMCID: PMC5908796
- DOI: 10.1038/s41467-018-03968-5
Culturing of female bladder bacteria reveals an interconnected urogenital microbiota
Abstract
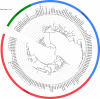
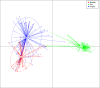
Metagenomic analyses have indicated that the female bladder harbors an indigenous microbiota. However, there are few cultured reference strains with sequenced genomes available for functional and experimental analyses. Here we isolate and genome-sequence 149 bacterial strains from catheterized urine of 77 women. This culture collection spans 78 species, representing approximately two thirds of the bacterial diversity within the sampled bladders, including Proteobacteria, Actinobacteria, and Firmicutes. Detailed genomic and functional comparison of the bladder microbiota to the gastrointestinal and vaginal microbiotas demonstrates similar vaginal and bladder microbiota, with functional capacities that are distinct from those observed in the gastrointestinal microbiota. Whole-genome phylogenetic analysis of bacterial strains isolated from the vagina and bladder in the same women identifies highly similar Escherichia coli, Streptococcus anginosus, Lactobacillus iners, and Lactobacillus crispatus, suggesting an interlinked female urogenital microbiota that is not only limited to pathogens but is also characteristic of health-associated commensals.
Conflict of interest statement
The authors declare no competing interests.
Figures

Comment in
-
What are bacteria doing in the bladder?Nat Rev Urol. 2018 Aug;15(8):469-470. doi: 10.1038/s41585-018-0032-y. Nat Rev Urol. 2018. PMID: 29844606 No abstract available.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

