cIAP1 regulates the EGFR/Snai2 axis in triple-negative breast cancer cells
- PMID: 29674627
- PMCID: PMC6262016
- DOI: 10.1038/s41418-018-0100-0
cIAP1 regulates the EGFR/Snai2 axis in triple-negative breast cancer cells
Abstract
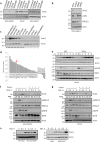
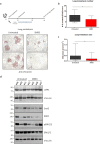
Inhibitor of apoptosis (IAP) proteins constitute a family of conserved molecules that regulate both apoptosis and receptor signaling. They are often deregulated in cancer cells and represent potential targets for therapy. In our work, we investigated the effect of IAP inhibition in vivo to identify novel downstream genes expressed in an IAP-dependent manner that could contribute to cancer aggressiveness. To this end, immunocompromised mice engrafted subcutaneously with the triple-negative breast cancer MDA-MB231 cell line were treated with SM83, a Smac mimetic that acts as a pan-IAP inhibitor, and tumor nodules were profiled for gene expression. SM83 reduced the expression of Snai2, an epithelial-to-mesenchymal transition factor often associated with increased stem-like properties and metastatic potential especially in breast cancer cells. By testing several breast cancer cell lines, we demonstrated that Snai2 downregulation prevents cell motility and that its expression is promoted by cIAP1. In fact, the chemical or genetic inhibition of cIAP1 blocked epidermal growth factor receptor (EGFR)-dependent activation of the mitogen-activated protein kinase (MAPK) pathway and caused the reduction of Snai2 transcription levels. In a number of breast cancer cell lines, cIAP1 depletion also resulted in a reduction of EGFR protein levels which derived from the decrease of its gene transcription, though, paradoxically, the silencing of cIAP1 promoted EGFR protein stability rather than its degradation. Finally, we provided evidence that IAP inhibition displays an anti-tumor and anti-metastasis effect in vivo. In conclusion, our work indicates that IAP-targeted therapy could contribute to EGFR inhibition and to the reduction of its downstream mediators. This approach could be particularly effective in tumors characterized by high levels of EGFR and Snai2, such as triple-negative breast cancer.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

