The molecular basis of JAK/STAT inhibition by SOCS1
- PMID: 29674694
- PMCID: PMC5908791
- DOI: 10.1038/s41467-018-04013-1
The molecular basis of JAK/STAT inhibition by SOCS1
Abstract
The SOCS family of proteins are negative-feedback inhibitors of signalling induced by cytokines that act via the JAK/STAT pathway. SOCS proteins can act as ubiquitin ligases by recruiting Cullin5 to ubiquitinate signalling components; however, SOCS1, the most potent member of the family, can also inhibit JAK directly. Here we determine the structural basis of both these modes of inhibition. Due to alterations within the SOCS box domain, SOCS1 has a compromised ability to recruit Cullin5; however, it is a direct, potent and selective inhibitor of JAK catalytic activity. The kinase inhibitory region of SOCS1 targets the substrate binding groove of JAK with high specificity and thereby blocks any subsequent phosphorylation. SOCS1 is a potent inhibitor of the interferon gamma (IFNγ) pathway, however, it does not bind the IFNγ receptor, making its mode-of-action distinct from SOCS3. These findings reveal the mechanism used by SOCS1 to inhibit signalling by inflammatory cytokines.
Conflict of interest statement
The authors declare no competing interests.
Figures
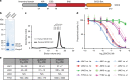

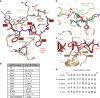
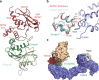
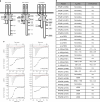

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

