Frequency of CYP2D6 Alleles Including Structural Variants in the United States
- PMID: 29674966
- PMCID: PMC5895772
- DOI: 10.3389/fphar.2018.00305
Frequency of CYP2D6 Alleles Including Structural Variants in the United States
Abstract
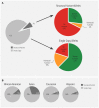
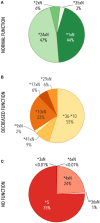
The CYP2D6 gene encodes an enzyme important in the metabolism of many commonly used medications. Variation in CYP2D6 is associated with inter-individual differences in medication response, and genetic testing is used to optimize medication therapy. This report describes a retrospective study of CYP2D6 allele frequencies in a large population of 104,509 de-identified patient samples across all regions of the United States (US). Thirty-seven unique CYP2D6 alleles including structural variants were identified. A majority of these alleles had frequencies which matched published frequency data from smaller studies, while eight had no previously published frequencies. Importantly, CYP2D6 structural variants were observed in 13.1% of individuals and accounted for 7% of the total variants observed. The majority of structural variants detected (73%) were decreased-function or no-function alleles. As such, structural variants were found in approximately one-third (30%) of CYP2D6 poor metabolizers in this study. This is the first CYP2D6 study to evaluate, with a consistent methodology, both structural variants and single copy alleles in a large US population, and the results suggest that structural variants have a substantial impact on CYP2D6 function.
Keywords: CYP2D6; copy number variation; cytochrome P450; drug metabolism; pharmacogenetics.
Figures




References
-
- (2016a). US Census Bureau: Divisions. Available online at: http://www2.census.gov/geo/pdfs/maps-data/maps/reference/us_regdiv.pdf
-
- (2016b). US Census Bureau: Ethnicity Percentages. Available online at: http://www.census.gov/quickfacts/table/PST045214/00
-
- Bell G. C., Caudle K. E., Whirl-Carrillo M., Gordon R. J., Hikino K., Prows C. A., et al. . (2017). Clinical Pharmacogenetics Implementation Consortium (CPIC) guideline for CYP2D6 genotype and use of ondansetron and tropisetron. Clin. Pharmacol. Ther. 102, 213–218. 10.1002/cpt.598 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

