Structure and Interaction Prediction in Prokaryotic RNA Biology
- PMID: 29676245
- PMCID: PMC11633574
- DOI: 10.1128/microbiolspec.RWR-0001-2017
Structure and Interaction Prediction in Prokaryotic RNA Biology
Abstract
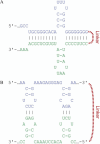
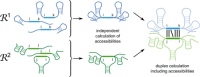
Many years of research in RNA biology have soundly established the importance of RNA-based regulation far beyond most early traditional presumptions. Importantly, the advances in "wet" laboratory techniques have produced unprecedented amounts of data that require efficient and precise computational analysis schemes and algorithms. Hence, many in silico methods that attempt topological and functional classification of novel putative RNA-based regulators are available. In this review, we technically outline thermodynamics-based standard RNA secondary structure and RNA-RNA interaction prediction approaches that have proven valuable to the RNA research community in the past and present. For these, we highlight their usability with a special focus on prokaryotic organisms and also briefly mention recent advances in whole-genome interactomics and how this may influence the field of predictive RNA research.
Figures





References
-
- Crick FH. 1966. Codon–anticodon pairing: the wobble hypothesis. J Mol Biol 19:548–555. - PubMed
-
- Gerber AP, Keller W. 1999. An adenosine deaminase that generates inosine at the wobble position of tRNAs. Science 286:1146–1149. - PubMed
-
- Watson JD, Crick FH. 1953. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature 171:737–738. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

