Mining TCGA database for genes of prognostic value in glioblastoma microenvironment
- PMID: 29676997
- PMCID: PMC5940130
- DOI: 10.18632/aging.101415
Mining TCGA database for genes of prognostic value in glioblastoma microenvironment
Abstract
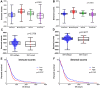
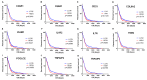
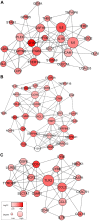
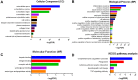
Glioblastoma (GBM) is one of the most deadly brain tumors. The convenient access to The Cancer Genome Atlas (TCGA) database allows for large-scale global gene expression profiling and database mining for potential correlation between genes and overall survival of a variety of malignancies including GBM. Previous reports have shown that tumor microenvironment cells and the extent of infiltrating immune and stromal cells in tumors contribute significantly to prognosis. Immune scores and stromal scores calculated based on the ESTIMATE algorithm could facilitate the quantification of the immune and stromal components in a tumor. To better understand the effects of genes involved in immune and stromal cells on prognosis, we categorized GBM cases in the TCGA database according to their immune/stromal scores into high and low score groups, and identified differentially expressed genes whose expression was significantly associated with prognosis in GBM patients. Functional enrichment analysis and protein-protein interaction networks further showed that these genes mainly participated in immune response, extracellular matrix, and cell adhesion. Finally, we validated these genes in an independent GBM cohort from the Chinese Glioma Genome Atlas (CGGA). Thus, we obtained a list of tumor microenvironment-related genes that predict poor outcomes in GBM patients.
Keywords: CGGA; TCGA; immune scores; overall survival; tumor microenvironment.
Conflict of interest statement
Figures


References
-
- Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn U, Curschmann J, Janzer RC, Ludwin SK, et al., and European Organisation for Research and Treatment of Cancer Brain Tumor and Radiotherapy Groups, and National Cancer Institute of Canada Clinical Trials Group. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005; 352:987–96. 10.1056/NEJMoa043330 - DOI - PubMed
-
- Perry JR, Laperriere N, O’Callaghan CJ, Brandes AA, Menten J, Phillips C, Fay M, Nishikawa R, Cairncross JG, Roa W, Osoba D, Rossiter JP, Sahgal A, et al., and Trial Investigators. Short-Course Radiation plus Temozolomide in Elderly Patients with Glioblastoma. N Engl J Med. 2017; 376:1027–37. 10.1056/NEJMoa1611977 - DOI - PubMed
-
- Verhaak RG, Hoadley KA, Purdom E, Wang V, Qi Y, Wilkerson MD, Miller CR, Ding L, Golub T, Mesirov JP, Alexe G, Lawrence M, O’Kelly M, et al., and Cancer Genome Atlas Research Network. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010; 17:98–110. 10.1016/j.ccr.2009.12.020 - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

