INDEL variation in the regulatory region of the major flowering time gene LanFTc1 is associated with vernalization response and flowering time in narrow-leafed lupin (Lupinus angustifolius L.)
- PMID: 29677403
- PMCID: PMC7379684
- DOI: 10.1111/pce.13320
INDEL variation in the regulatory region of the major flowering time gene LanFTc1 is associated with vernalization response and flowering time in narrow-leafed lupin (Lupinus angustifolius L.)
Abstract
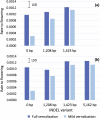
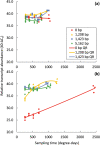
Narrow-leafed lupin (Lupinus angustifolius L.) cultivation was transformed by 2 dominant vernalization-insensitive, early flowering time loci known as Ku and Julius (Jul), which allowed expansion into shorter season environments. However, reliance on these loci has limited genetic and phenotypic diversity for environmental adaptation in cultivated lupin. We recently predicted that a 1,423-bp deletion in the cis-regulatory region of LanFTc1, a FLOWERING LOCUS T (FT) homologue, derepressed expression of LanFTc1 and was the underlying cause of the Ku phenotype. Here, we surveyed diverse germplasm for LanFTc1 cis-regulatory variation and identified 2 further deletions of 1,208 and 5,162 bp in the 5' regulatory region, which overlap the 1,423-bp deletion. Additionally, we confirmed that no other polymorphisms were perfectly associated with vernalization responsiveness. Phenotyping and gene expression analyses revealed that Jul accessions possessed the 5,162-bp deletion and that the Jul and Ku deletions were equally capable of removing vernalization requirement and up-regulating gene expression. The 1,208-bp deletion was associated with intermediate phenology, vernalization responsiveness, and gene expression and therefore may be useful for expanding agronomic adaptation of lupin. This insertion/deletion series may also help resolve how the vernalization response is mediated at the molecular level in legumes.
Keywords: FLOWERING LOCUS T (FT); cis-regulation; insertion/deletion (INDEL); variant series.
© 2018 The Authors Plant, Cell & Environment Published by John Wiley & Sons Ltd.
Figures


References
-
- Barker, S. J. , Si, P. , Hodgson, L. , Ferguson‐Hunt, M. , Khentry, Y. , Krishnamurthy, P. , … Erskine, W. (2016). Regeneration selection improves transformation efficiency in narrow‐leaf lupin. Plant Cell Tissue and Organ Culture, 126, 219–228.
-
- Berger, J. , Buirchell, B. , Palta, J. , Luckett, D. , Ludwig, C. , & Shrestha, D. (2008). G x E analysis of narrow‐leafed lupin historical trials indicates little specific adaptation among Australian cultivars In Palta J. A., & Berger J. D. (Eds.), Lupins for Health and Wealth (pp. 317–320. Proceedings of the 12th International Lupin Conference, 14‐18 September 2008, Fremantle, Western Australia). Canterbury, New Zealand: International Lupin Association.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

