Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing
- PMID: 29677493
- PMCID: PMC5916843
- DOI: 10.1016/j.molcel.2018.03.015
Polyamine Control of Translation Elongation Regulates Start Site Selection on Antizyme Inhibitor mRNA via Ribosome Queuing
Abstract
Translation initiation is typically restricted to AUG codons, and scanning eukaryotic ribosomes inefficiently recognize near-cognate codons. We show that queuing of scanning ribosomes behind a paused elongating ribosome promotes initiation at upstream weak start sites. Ribosomal profiling reveals polyamine-dependent pausing of elongating ribosomes on a conserved Pro-Pro-Trp (PPW) motif in an inhibitory non-AUG-initiated upstream conserved coding region (uCC) of the antizyme inhibitor 1 (AZIN1) mRNA, encoding a regulator of cellular polyamine synthesis. Mutation of the PPW motif impairs initiation at the uCC's upstream near-cognate AUU start site and derepresses AZIN1 synthesis, whereas substitution of alternate elongation pause sequences restores uCC translation. Impairing ribosome loading reduces uCC translation and paradoxically derepresses AZIN1 synthesis. Finally, we identify the translation factor eIF5A as a sensor and effector for polyamine control of uCC translation. We propose that stalling of elongating ribosomes triggers queuing of scanning ribosomes and promotes initiation by positioning a ribosome near the start codon.
Keywords: AZIN1; antizyme inhibitor; eIF5A; near cognate; polyamines; ribosome; spermidine.
Published by Elsevier Inc.
Conflict of interest statement
G.L. is a founder and shareholder of RiboMaps Ltd, a company providing ribosome profiling as a service. As the findings reported here are supported by ribosome profiling, its publication may increase the attractiveness of this technology and indirectly benefit the company.
Figures

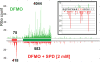

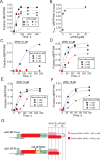
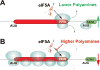
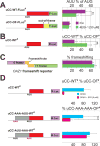
Comment in
-
The Traffic Jam: Polyamine Prevalence Pauses Protein Production.Mol Cell. 2018 Apr 19;70(2):191-192. doi: 10.1016/j.molcel.2018.04.005. Mol Cell. 2018. PMID: 29677487
Similar articles
-
Translational autoregulation of the S. cerevisiae high-affinity polyamine transporter Hol1.Mol Cell. 2021 Oct 7;81(19):3904-3918.e6. doi: 10.1016/j.molcel.2021.07.020. Epub 2021 Aug 9. Mol Cell. 2021. PMID: 34375581 Free PMC article.
-
Suppression of Ribosomal Pausing by eIF5A Is Necessary to Maintain the Fidelity of Start Codon Selection.Cell Rep. 2019 Dec 3;29(10):3134-3146.e6. doi: 10.1016/j.celrep.2019.10.129. Cell Rep. 2019. PMID: 31801078 Free PMC article.
-
eIF5A Functions Globally in Translation Elongation and Termination.Mol Cell. 2017 Apr 20;66(2):194-205.e5. doi: 10.1016/j.molcel.2017.03.003. Epub 2017 Apr 6. Mol Cell. 2017. PMID: 28392174 Free PMC article.
-
Roles of polyamines in translation.J Biol Chem. 2018 Nov 30;293(48):18719-18729. doi: 10.1074/jbc.TM118.003338. Epub 2018 Oct 15. J Biol Chem. 2018. PMID: 30323064 Free PMC article. Review.
-
eIF5A and EF-P: two unique translation factors are now traveling the same road.Wiley Interdiscip Rev RNA. 2014 Mar-Apr;5(2):209-22. doi: 10.1002/wrna.1211. Epub 2014 Jan 8. Wiley Interdiscip Rev RNA. 2014. PMID: 24402910 Review.
Cited by
-
Polyamine Homeostasis in Development and Disease.Med Sci (Basel). 2021 May 13;9(2):28. doi: 10.3390/medsci9020028. Med Sci (Basel). 2021. PMID: 34068137 Free PMC article. Review.
-
Polyamine Depletion Inhibits Bunyavirus Infection via Generation of Noninfectious Interfering Virions.J Virol. 2019 Jun 28;93(14):e00530-19. doi: 10.1128/JVI.00530-19. Print 2019 Jul 15. J Virol. 2019. PMID: 31043534 Free PMC article.
-
Ribosome decision graphs for the representation of eukaryotic RNA translation complexity.Genome Res. 2024 May 15;34(4):530-538. doi: 10.1101/gr.278810.123. Genome Res. 2024. PMID: 38719470 Free PMC article.
-
From Recoding to Peptides for MHC Class I Immune Display: Enriching Viral Expression, Virus Vulnerability and Virus Evasion.Viruses. 2021 Jun 27;13(7):1251. doi: 10.3390/v13071251. Viruses. 2021. PMID: 34199077 Free PMC article. Review.
-
Determinants of genome-wide distribution and evolution of uORFs in eukaryotes.Nat Commun. 2021 Feb 17;12(1):1076. doi: 10.1038/s41467-021-21394-y. Nat Commun. 2021. PMID: 33597535 Free PMC article.
References
-
- Anand M, Balar B, Ulloque R, Gross SR, Kinzy TG. Domain and nucleotide dependence of the interaction between Saccharomyces cerevisiae translation elongation factors 3 and 1A. J Biol Chem. 2006;281:32318–32326. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

