The prevalence of molecular markers of drug resistance in Plasmodium vivax from the border regions of Thailand in 2008 and 2014
- PMID: 29677637
- PMCID: PMC6039358
- DOI: 10.1016/j.ijpddr.2018.04.003
The prevalence of molecular markers of drug resistance in Plasmodium vivax from the border regions of Thailand in 2008 and 2014
Abstract
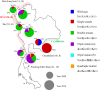
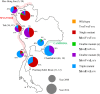
The prevalence of Plasmodium vivax is increasing in the border regions of Thailand; one potential problem confounding the control of malaria in these regions is the emergence and spread of drug resistance. The aim of this study was to determine the genetic diversity in genes potentially linked to drug resistance in P. vivax parasites isolated from four different border regions of Thailand; Thai-Myanmar (Tak, Mae Hong Son and Prachuap Khiri Khan Provinces), and Thai-Cambodian borders (Chanthaburi Province). Isolates were collected from 345 P. vivax patients in 2008 and 2014, and parasite DNA extracted and subjected to nucleotide sequencing at five putative drug-resistance loci (Pvdhfr, Pvdhps, Pvmdr1, Pvcrt-o and Pvk12). The prevalence of mutations in Pvdhfr, Pvdhps and Pvmdr1 were markedly different between the Thai-Myanmar and Thai-Cambodian border areas and also varied between sampling times. All isolates carried the Pvdhfr (58R and 117N/T) mutation, however, whereas the quadruple mutant allele (I57R58M61T117) was the most prevalent (69.6%) in the Thai-Myanmar border region, the double mutant allele (F57R58T61N117) was at fixation on the Thai-Cambodian border (100%). The most prevalent genotypes of Pvdhps and Pvmdr1 were the double mutant (S382G383K512G553) (65.1%) and single mutant (M958Y976F1076) (46.5%) alleles, respectively on the Thai-Myanmar border while the single Pvdhps mutant (S382G383K512A553) (52.7%) and the triple Pvmdr1 mutant (M958F976L1076) (81%) alleles were dominant on the Thai-Cambodian border. No mutations were observed in the Pvcrt-o gene in either region. Novel mutations in the Pvk12 gene, the P. vivax orthologue of PfK13, linked to artemisinin resistance in Plasmodium falciparum, were observed with three nonsynonymous and three synonymous mutations in six isolates (3.3%).
Keywords: Antimalarial drugs; Drug-resistant mutations; Genetic diversity; Plasmodium vivax.
Copyright © 2018 The Authors. Published by Elsevier Ltd.. All rights reserved.
Figures



References
-
- Ariey F., Witkowski B., Amaratunga C., Beghain J., Langlois A.C., Khim N., Kim S., Duru V., Bouchier C., Ma L., Lim P., Leang R., Duong S., Sreng S., Suon S., Chuor C.M., Bout D.M., Menard S., Rogers W.O., Genton B., Fandeur T., Miotto O., Ringwald P., Le Bras J., Berry A., Barale J.C., Fairhurst R.M., Benoit-Vical F., Mercereau-Puijalon O., Menard D. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature. 2014;505:50–55. - PMC - PubMed
-
- Ashley E.A., Dhorda M., Fairhurst R.M., Amaratunga C., Lim P., Suon S., Sreng S., Anderson J.M., Mao S., Sam B., Sopha C., Chuor C.M., Nguon C., Sovannaroth S., Pukrittayakamee S., Jittamala P., Chotivanich K., Chutasmit K., Suchatsoonthorn C., Runcharoen R., Hien T.T., Thuy-Nhien N.T., Thanh N.V., Phu N.H., Htut Y., Han K.T., Aye K.H., Mokuolu O.A., Olaosebikan R.R., Folaranmi O.O., Mayxay M., Khanthavong M., Hongvanthong B., Newton P.N., Onyamboko M.A., Fanello C.I., Tshefu A.K., Mishra N., Valecha N., Phyo A.P., Nosten F., Yi P., Tripura R., Borrmann S., Bashraheil M., Peshu J., Faiz M.A., Ghose A., Hossain M.A., Samad R., Rahman M.R., Hasan M.M., Islam A., Miotto O., Amato R., MacInnis B., Stalker J., Kwiatkowski D.P., Bozdech Z., Jeeyapant A., Cheah P.Y., Sakulthaew T., Chalk J., Intharabut B., Silamut K., Lee S.J., Vihokhern B., Kunasol C., Imwong M., Tarning J., Taylor W.J., Yeung S., Woodrow C.J., Flegg J.A., Das D., Smith J., Venkatesan M., Plowe C.V., Stepniewska K., Guerin P.J., Dondorp A.M., Day N.P., White N.J., Tracking Resistance to Artemisinin, C Spread of artemisinin resistance in Plasmodium falciparum malaria. N. Engl. J. Med. 2014;371:411–423. - PMC - PubMed
-
- Auliff A., Wilson D.W., Russell B., Gao Q., Chen N., Anh le N., Maguire J., Bell D., O'Neil M.T., Cheng Q. Amino acid mutations in Plasmodium vivax DHFR and DHPS from several geographical regions and susceptibility to antifolate drugs. Am. J. Trop. Med. Hyg. 2006;75:617–621. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

