Regulation of Cell-to-Cell Communication and Cell Wall Integrity by a Network of MAP Kinase Pathways and Transcription Factors in Neurospora crassa
- PMID: 29678830
- PMCID: PMC5972422
- DOI: 10.1534/genetics.118.300904
Regulation of Cell-to-Cell Communication and Cell Wall Integrity by a Network of MAP Kinase Pathways and Transcription Factors in Neurospora crassa
Abstract
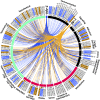
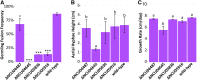
Maintenance of cell integrity and cell-to-cell communication are fundamental biological processes. Filamentous fungi, such as Neurospora crassa, depend on communication to locate compatible cells, coordinate cell fusion, and establish a robust hyphal network. Two MAP kinase (MAPK) pathways are essential for communication and cell fusion in N. crassa: the cell wall integrity/MAK-1 pathway and the MAK-2 (signal response) pathway. Previous studies have demonstrated several points of cross-talk between the MAK-1 and MAK-2 pathways, which is likely necessary for coordinating chemotropic growth toward an extracellular signal, and then mediating cell fusion. Canonical MAPK pathways begin with signal reception and end with a transcriptional response. Two transcription factors, ADV-1 and PP-1, are essential for communication and cell fusion. PP-1 is the conserved target of MAK-2, but it is unclear what targets ADV-1. We did RNA sequencing on Δadv-1, Δpp-1, and wild-type cells and found that ADV-1 and PP-1 have a shared regulon including many genes required for communication, cell fusion, growth, development, and stress response. We identified ADV-1 and PP-1 binding sites across the genome by adapting the in vitro method of DNA-affinity purification sequencing for N. crassa To elucidate the regulatory network, we misexpressed each transcription factor in each upstream MAPK deletion mutant. Misexpression of adv-1 was sufficient to fully suppress the phenotype of the Δpp-1 mutant and partially suppress the phenotype of the Δmak-1 mutant. Collectively, our data demonstrate that the MAK-1/ADV-1 and MAK-2/PP-1 pathways form a tight regulatory network that maintains cell integrity and mediates communication and cell fusion.
Keywords: DAP-seq; MAP kinase; Neurospora crassa; cell fusion; cell-to-cell communication; regulatory networks.
Copyright © 2018 by the Genetics Society of America.
Figures







Similar articles
-
Transcriptional profiling of Neurospora crassa Δmak-2 reveals that mitogen-activated protein kinase MAK-2 participates in the phosphate signaling pathway.Fungal Genet Biol. 2013 Nov;60:140-9. doi: 10.1016/j.fgb.2013.05.007. Epub 2013 Jun 1. Fungal Genet Biol. 2013. PMID: 23733042
-
Involvement of MAK-1 and MAK-2 MAP kinases in cell wall integrity in Neurospora crassa.Biosci Biotechnol Biochem. 2016 Sep;80(9):1843-52. doi: 10.1080/09168451.2016.1189321. Epub 2016 Jun 7. Biosci Biotechnol Biochem. 2016. PMID: 27268441
-
HAM-5 functions as a MAP kinase scaffold during cell fusion in Neurospora crassa.PLoS Genet. 2014 Nov 20;10(11):e1004783. doi: 10.1371/journal.pgen.1004783. eCollection 2014 Nov. PLoS Genet. 2014. PMID: 25412208 Free PMC article.
-
Cell fusion in Neurospora crassa.Curr Opin Microbiol. 2015 Dec;28:53-9. doi: 10.1016/j.mib.2015.08.002. Epub 2015 Sep 3. Curr Opin Microbiol. 2015. PMID: 26340439 Review.
-
White collar proteins: PASsing the light signal in Neurospora crassa.Trends Microbiol. 1997 Nov;5(11):458-62. doi: 10.1016/S0966-842X(97)01144-X. Trends Microbiol. 1997. PMID: 9402704 Review.
Cited by
-
Synchronization of oscillatory growth prepares fungal hyphae for fusion.Elife. 2023 Aug 21;12:e83310. doi: 10.7554/eLife.83310. Elife. 2023. PMID: 37602797 Free PMC article.
-
Broad Substrate-Specific Phosphorylation Events Are Associated With the Initial Stage of Plant Cell Wall Recognition in Neurospora crassa.Front Microbiol. 2019 Nov 1;10:2317. doi: 10.3389/fmicb.2019.02317. eCollection 2019. Front Microbiol. 2019. PMID: 31736884 Free PMC article.
-
Quorum sensing-mediated inter-specific conidial anastomosis tube fusion between Colletotrichum gloeosporioides and C. siamense.IMA Fungus. 2021 Apr 1;12(1):7. doi: 10.1186/s43008-021-00058-y. IMA Fungus. 2021. PMID: 33789776 Free PMC article.
-
AoSte12 Is Required for Mycelial Development, Conidiation, Trap Morphogenesis, and Secondary Metabolism by Regulating Hyphal Fusion in Nematode-Trapping Fungus Arthrobotrys oligospora.Microbiol Spectr. 2023 Feb 14;11(2):e0395722. doi: 10.1128/spectrum.03957-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36786575 Free PMC article.
-
Integration of Self and Non-self Recognition Modulates Asexual Cell-to-Cell Communication in Neurospora crassa.Genetics. 2019 Apr;211(4):1255-1267. doi: 10.1534/genetics.118.301780. Epub 2019 Feb 4. Genetics. 2019. PMID: 30718271 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

