GSyellow, a Multifaceted Tag for Functional Protein Analysis in Monocot and Dicot Plants
- PMID: 29678859
- PMCID: PMC6001315
- DOI: 10.1104/pp.18.00175
GSyellow, a Multifaceted Tag for Functional Protein Analysis in Monocot and Dicot Plants
Abstract
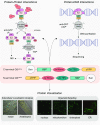
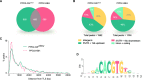
The ability to tag proteins has boosted the emergence of generic molecular methods for protein functional analysis. Fluorescent protein tags are used to visualize protein localization, and affinity tags enable the mapping of molecular interactions by, for example, tandem affinity purification or chromatin immunoprecipitation. To apply these widely used molecular techniques on a single transgenic plant line, we developed a multifunctional tandem affinity purification tag, named GSyellow, which combines the streptavidin-binding peptide tag with citrine yellow fluorescent protein. We demonstrated the versatility of the GSyellow tag in the dicot Arabidopsis (Arabidopsis thaliana) using a set of benchmark proteins. For proof of concept in monocots, we assessed the localization and dynamic interaction profile of the leaf growth regulator ANGUSTIFOLIA3 (AN3), fused to the GSyellow tag, along the growth zone of the maize (Zea mays) leaf. To further explore the function of ZmAN3, we mapped its DNA-binding landscape in the growth zone of the maize leaf through chromatin immunoprecipitation sequencing. Comparison with AN3 target genes mapped in the developing maize tassel or in Arabidopsis cell cultures revealed strong conservation of AN3 target genes between different maize tissues and across monocots and dicots, respectively. In conclusion, the GSyellow tag offers a powerful molecular tool for distinct types of protein functional analyses in dicots and monocots. As this approach involves transforming a single construct, it is likely to accelerate both basic and translational plant research.
© 2018 American Society of Plant Biologists. All rights reserved.
Figures




Similar articles
-
Distinct functional properties of isoamylase-type starch debranching enzymes in monocot and dicot leaves.Plant Physiol. 2013 Nov;163(3):1363-75. doi: 10.1104/pp.113.225565. Epub 2013 Sep 11. Plant Physiol. 2013. PMID: 24027240 Free PMC article.
-
Dynamic Changes in ANGUSTIFOLIA3 Complex Composition Reveal a Growth Regulatory Mechanism in the Maize Leaf.Plant Cell. 2015 Jun;27(6):1605-19. doi: 10.1105/tpc.15.00269. Epub 2015 Jun 2. Plant Cell. 2015. PMID: 26036253 Free PMC article.
-
Developmental changes in abundance of the VSPbeta protein following nuclear transformation of maize with the soybean vspbeta cDNA.BMC Plant Biol. 2005 Mar 2;5:3. doi: 10.1186/1471-2229-5-3. BMC Plant Biol. 2005. PMID: 15743517 Free PMC article.
-
Methods for generation and analysis of fluorescent protein-tagged maize lines.Methods Mol Biol. 2009;526:71-89. doi: 10.1007/978-1-59745-494-0_6. Methods Mol Biol. 2009. PMID: 19378001
-
Fluorescent protein marker lines in maize: generation and applications.Int J Dev Biol. 2013;57(6-8):535-43. doi: 10.1387/ijdb.130240qw. Int J Dev Biol. 2013. PMID: 24166436 Review.
Cited by
-
From Affinity to Proximity Techniques to Investigate Protein Complexes in Plants.Int J Mol Sci. 2021 Jul 1;22(13):7101. doi: 10.3390/ijms22137101. Int J Mol Sci. 2021. PMID: 34281155 Free PMC article. Review.
-
The membrane-localized protein kinase MAP4K4/TOT3 regulates thermomorphogenesis.Nat Commun. 2021 May 14;12(1):2842. doi: 10.1038/s41467-021-23112-0. Nat Commun. 2021. PMID: 33990595 Free PMC article.
-
Mapping the plant proteome: tools for surveying coordinating pathways.Emerg Top Life Sci. 2021 May 21;5(2):203-220. doi: 10.1042/ETLS20200270. Emerg Top Life Sci. 2021. PMID: 33620075 Free PMC article. Review.
-
SAMBA controls cell division rate during maize development.Plant Physiol. 2022 Jan 20;188(1):411-424. doi: 10.1093/plphys/kiab514. Plant Physiol. 2022. PMID: 34791456 Free PMC article.
-
PIF7 controls leaf cell proliferation through an AN3 substitution repression mechanism.Proc Natl Acad Sci U S A. 2022 Feb 1;119(5):e2115682119. doi: 10.1073/pnas.2115682119. Proc Natl Acad Sci U S A. 2022. PMID: 35086930 Free PMC article.
References
-
- Bürckstümmer T, Bennett KL, Preradovic A, Schütze G, Hantschel O, Superti-Furga G, Bauch A (2006) An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat Methods 3: 1013–1019 - PubMed
-
- Cheeseman IM, Desai A (2005) A combined approach for the localization and tandem affinity purification of protein complexes from metazoans. Sci STKE 2005: pl1. - PubMed
-
- Coussens G, Aesaert S, Verelst W, Demeulenaere M, De Buck S, Njuguna E, Inzé D, Van Lijsebettens M (2012) Brachypodium distachyon promoters as efficient building blocks for transgenic research in maize. J Exp Bot 63: 4263–4273 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

