Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli
- PMID: 29678883
- PMCID: PMC5995498
- DOI: 10.1074/jbc.RA117.000300
Ribosome maturation by the endoribonuclease YbeY stabilizes a type 3 secretion system transcript required for virulence of enterohemorrhagic Escherichia coli
Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a significant human pathogen that colonizes humans and its reservoir host, cattle. Colonization requires the expression of a type 3 secretion (T3S) system that injects a mixture of effector proteins into host cells to promote bacterial attachment and disease progression. The T3S system is tightly regulated by a complex network of transcriptional and post-transcriptional regulators. Using transposon mutagenesis, here we identified the ybeZYX-Int operon as being required for normal T3S levels. Deletion analyses localized the regulation to the endoribonuclease YbeY, previously linked to 16S rRNA maturation and small RNA (sRNA) function. Loss of ybeY in EHEC had pleiotropic effects on EHEC cells, including reduced motility and growth and cold sensitivity. Using UV cross-linking and RNA-Seq (CRAC) analysis, we identified YbeY-binding sites throughout the transcriptome and discovered specific binding of YbeY to the "neck" and "beak" regions of 16S rRNA but identified no significant association of YbeY with sRNA, suggesting that YbeY modulates T3S by depleting mature ribosomes. In E. coli, translation is strongly linked to mRNA stabilization, and subinhibitory concentrations of the translation-initiation inhibitor kasugamycin provoked rapid degradation of a polycistronic mRNA encoding needle filament and needle tip proteins of the T3S system. We conclude that T3S is particularly sensitive to depletion of initiating ribosomes, explaining the inhibition of T3S in the ΔybeY strain. Accessory virulence transcripts may be preferentially degraded in cells with reduced translational capacity, potentially reflecting prioritization in protein production.
Keywords: RNA degradation; YbeY; anti-virulence; antibiotics; bacterial pathogenesis; enterohemorrhagic E. coli; gene regulation; ketolide; post-transcriptional regulation; precursor ribosomal RNA (pre-rRNA); ribonuclease; ribosomal RNA processing (rRNA processing); ribosomal ribonucleic acid (rRNA) (ribosomal RNA); ribosome assembly; small RNA; type 3 secretion; type III secretion; virulence factor.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures

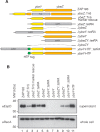
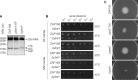

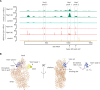

References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

