Role of the novel endoribonuclease SLFN14 and its disease-causing mutations in ribosomal degradation
- PMID: 29678925
- PMCID: PMC6004054
- DOI: 10.1261/rna.066415.118
Role of the novel endoribonuclease SLFN14 and its disease-causing mutations in ribosomal degradation
Abstract
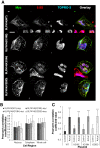
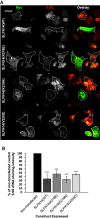
Platelets are anucleate and mostly ribosome-free cells within the bloodstream, derived from megakaryocytes within bone marrow and crucial for cessation of bleeding at sites of injury. Inherited thrombocytopenias are a group of disorders characterized by a low platelet count and are frequently associated with excessive bleeding. SLFN14 is one of the most recently discovered genes linked to inherited thrombocytopenia where several heterozygous missense mutations in SLFN14 were identified to cause defective megakaryocyte maturation and platelet dysfunction. Yet, SLFN14 was recently described as a ribosome-associated protein resulting in rRNA and ribosome-bound mRNA degradation in rabbit reticulocytes. To unveil the cellular function of SLFN14 and the link between SLFN14 and thrombocytopenia, we examined SLFN14 (WT/mutants) in in vitro models. Here, we show that all SLFN14 variants colocalize with ribosomes and mediate rRNA endonucleolytic degradation. Compared to SLFN14 WT, expression of mutants is dramatically reduced as a result of post-translational degradation due to partial misfolding of the protein. Moreover, all SLFN14 variants tend to form oligomers. These findings could explain the dominant negative effect of heterozygous mutation on SLFN14 expression in patients' platelets. Overall, we suggest that SLFN14 could be involved in ribosome degradation during platelet formation and maturation.
Keywords: RNA degradation; SLFN14; endoribonuclease; platelet; ribonuclease; thrombocytopenia.
© 2018 Fletcher et al.; Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures


References
-
- Brady G, Boggan L, Bowie A, O'Neill LAJ. 2005. Schlafen-1 causes a cell cycle arrest by inhibiting induction of cyclin D1. J Biol Chem 280: 30723–30734. - PubMed
-
- Cox TM, Cachón-González MB. 2012. The cellular pathology of lysosomal diseases. J Pathol 226: 241–254. - PubMed
-
- Favier R, Raslova H. 2015. Progress in understanding the diagnosis and molecular genetics of macrothrombocytopenias. Br J Haematol 170: 626–639. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- PG/13/36/30275/BHF_/British Heart Foundation/United Kingdom
- R01 GM097014/GM/NIGMS NIH HHS/United States
- PG/16/103/32650/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- PG/13/36/30275 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- PG/16/103/32650/BHF_/British Heart Foundation/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
