Self-replication of DNA by its encoded proteins in liposome-based synthetic cells
- PMID: 29679002
- PMCID: PMC5910420
- DOI: 10.1038/s41467-018-03926-1
Self-replication of DNA by its encoded proteins in liposome-based synthetic cells
Abstract
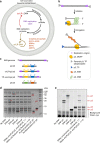
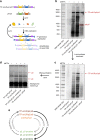
Replication of DNA-encoded information and its conversion into functional proteins are universal properties of life. In an effort toward the construction of a synthetic minimal cell, we implement here the DNA replication machinery of the Φ29 virus in a cell-free gene expression system. Amplification of a linear DNA template by self-encoded, de novo synthesized Φ29 proteins is demonstrated. Complete information transfer is confirmed as the copied DNA can serve as a functional template for gene expression, which can be seen as an autocatalytic DNA replication cycle. These results show how the central dogma of molecular biology can be reconstituted and form a cycle in vitro. Finally, coupled DNA replication and gene expression is compartmentalized inside phospholipid vesicles providing the chassis for evolving functions in a prospective synthetic cell relying on the extant biology.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

